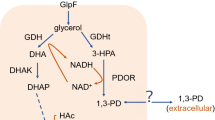
Abstract
Understanding the transport mechanism of 1,3-propanediol (1,3-PD) is of critical importance to do further research on gene regulation. Due to the lack of intracellular information, on the basis of enzyme-catalytic system, using biological robustness as performance index, we present a system identification model to infer the most possible transport mechanism of 1,3-PD, in which the performance index consists of the relative error of the extracellular substance concentrations and biological robustness of the intracellular substance concentrations. We will not use a Boolean framework but prefer a model description based on ordinary differential equations. Among other advantages, this also facilitates the robustness analysis, which is the main goal of this paper. An algorithm is constructed to seek the solution of the identification model. Numerical results show that the most possible transport way is active transport coupled with passive diffusion.






Similar content being viewed by others
References
Bibel H, Memzel K, Zeng AP, Deckwer WD (1999) Microbial production of 1,3-propanediol. Appl Microbiol Biotechnol 52:289–297
Xiu ZL (2000) Research progress on the production of 1,3-propanediol by fermentation. Microbiology 27:300–302
Xu GX (2010) Robust control of continuous bioprocesses. Math Probl Eng 2010:Article ID 627035
Xu GX (2012) Bi-objective optimization of biochemical systems by linear programming. Appl Math Comput 218:7562–7572
Tian Y, Sun KB, Kasperski A, Chen LS (2010) Studies on the dynamics of a continuous bioprocess with impulsive state feedback control. Chem Eng J 157:558–567
Liu CY, Gong ZH, Feng EM, Yin HC (2009) Modelling and optimal control for nonlinear multistage dynamical system of microbial fed-batch culture. J Ind Manag Optim 5(4):835–850
Gao CX, Feng EM, Wang ZT, Xiu ZL (2005) Nonlinear dynamical systems of bio-dissimilation of glycerol to 1,3-propanediol and their optimal controls. J Ind Manag Optim 1:377–388
Wang G, Feng EM, Xiu ZL (2008) Modeling and parameter identification of microbial bioconversion in fed-batch cultures. J Process Control 18(5):458–464
Wang L, Xiu ZL, Gong ZH, Feng EM (2012) Modeling and parameter identification for multistage simulation of microbial bioconversion in batch culture. Int J Biomath. doi:10.1142/S179352451100174X
Wang HY, Feng EM, Xiu ZL (2008) Optimality condition of the nonlinear impulsive system in fed-batch fermentation. Nonlinear Anal Theory Methods Appl 68(1):12–23
Li XH, Guo JJ, Feng EM, Xiu ZL (2010) Discrete optimal control model and bound error for microbial continuous fermentation. Nonlinear Anal Real World Appl 11(1):131–138
Wang L, Xiu ZL, Zhang YD, Feng EM (2011) Optimal control for multistage nonlinear dynamic system of microbial bioconversion in batch culture, J Appl Math 2011:Article ID 624516
Wang L (2012) Modelling and regularity of nonlinear impulsive switching dynamical system in fed-batch culture, abstract and applied analysis 2012:Article ID 295627
Zhang QR, Teng H, Wu ZN, Xiu ZL (2008) Process dynamic modeling based on metabolic network of glycerol bioconversion to 1,3-propanediol. J Biotechnol 136S:S22–S71
Tian Y, Sun KB, Kasperski A, Chen LS (2010) Nonlinear modelling and qualitative analysis of a real chemostat with pulse feeding. Discret Dyn Nat Soc 2010:Article ID 640594
Ashoori A, Moshiri B, Khaki-Sedigh A, Bakhtiari MR (2009) Optimal control of a nonlinear fed-batch fermentation process using model predictive approach. J Process Control 19:1162–1173
Gong ZH, Liu CY, Feng EM, Zhang QR (2010) Computational method for Infer objective function of glycerol metabolism in klebsiella pneumonia basing on bilevel programming. J Syst Sci Complex 23(2):334–342
Sun YQ, Qi WT, Teng H, Xiu ZL, Zeng AP (2008) Mathematical modeling of glycerol fermentation by klebsiella pneumoniae: concerning enzyme catalytic reductive pathway and transport of glycerol and 1,3-propanediol across cell membrane. Biochem Eng J 38:22–32
Zhang Q, Teng H, Sun Y, Xiu Z, Zeng A (2008) Metabolic flux and robustness analysis of glycerol metabolism in Klebsiella pneumonia. Bioprocess Biosyst Eng 31:127–135
Ye J, Feng E et al (2009) Modeling and robustness analysis of biochemical networks of glycerol metabolism by Klebsiella Pneumoniae. Lecture Notes Inst Comput Sci Soc Informa Telecommun Eng 4(1):446–457
Wang J et al (2011) Complex metabolic network of glycerol fermentation by Klebsiella pneumoniae and its system identification via biological robustness. Nonlinear Anal Hybrid Syst 5:102–112
Wang J et al (2011) Modeling and identification of a nonlinear hybrid dynamical system in batch fermentation of glycerol. Math Comput Model 54:618–624
Yan H, Zhang X, Ye J, Feng E (2012) Identification and robustness analysis of nonlinear hybrid dynamical system concerning glycerol transport mechanism. Comput Chem Eng 40:171–180
Zhai J, Ye J et al (2011) Pathway identification using parallel optimization for a complex metabolic system in microbial continuous culture. Nonlinear Anal Real World Appl 12(5):2730–2741
Zeng AP, Biebl H (2002) Bulk chemicals from biotechnology: the case of 1.3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74:239–259
Kitano Hiroaki (2004) Biological robustness. Nat Rev Genet 5:826–837
Stelling Jorg, Sauer Uwe, Szallasi Zoltan (2004) Robustness of cellular functions. Cell 118:675–685
Sleuer Ralf (2007) Computational approaches to the topology, stability and dynamics of metabolic networks. Phytochemistry 68:2139–2151
Sun J, van den Heuvel J, Soucaille P, Qu Y, Zeng AP (2003) Comparative genomic analysis of dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol Prog 19:263–272
Wang SY, Feng EM (2010) Stability of nonlinear microbial bioconversion system concerning glycerol’s active transport and 1, 3-PD’s passive transport. Nonlinear Analysis: Real World Applications 11:3501–3511
Torres N’estor V, Voit Eberhard O (2002) Pathway analysis and optimization in metabolic engineering. Cambridge University Press, Cambridge
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (No. DUT12LK27), the National Natural Science Foundation of China (Grant Nos. 10671126, 10871033 and 11171050) and the Natural Science Foundation for the Youth of China (No. 11001153).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, L. Determining the transport mechanism of an enzyme-catalytic complex metabolic network based on biological robustness. Bioprocess Biosyst Eng 36, 433–441 (2013). https://doi.org/10.1007/s00449-012-0800-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-012-0800-7