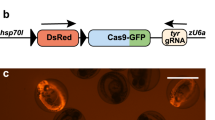
Abstract
New genome-editing approaches, such as the CRISPR/Cas system, have opened up great opportunities to insert or delete genes at targeted loci and have revolutionized genetics in model organisms like the zebrafish. The Cre-loxp recombination system is widely used to activate or inactivate genes with high spatial and temporal specificity. Using a CRISPR/Cas9-mediated knock-in strategy, we inserted a zebrafish codon-optimized CreERT2 transgene at the otx2 gene locus to generate a conditional Cre-driver line. We chose otx2 as it is a patterning gene of the anterior neural plate that is expressed during early development. By knocking in CreERT2 upstream of the endogenous ATG of otx2, we utilized this gene’s native promoter and enhancer elements to perfectly match CreERT2 and endogenous otx2 expression patterns. Next, by combining this novel driver line with a Cre-dependent reporter line, we show that only in the presence of tamoxifen can efficient Cre-loxp-mediated recombination be achieved in the anterior neural plate-derived tissues like the telencephalon, the eye and the optic tectum. Our results imply that the otx2:CreERT2 transgenic fish will be a valuable tool for lineage tracing and conditional mutant studies in larval and adult zebrafish.





Similar content being viewed by others
References
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Lun K, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA et al (1996) Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development 123:179–190
Brand M, Granato M, Nüsslein-Volhard C (2002) Keeping and raising zebrafish. In: Nüsslein-Volhard C, Dahm R (eds) Zebrafish: a practical approach. Oxford University Press, Oxford, pp 7–37
Chekuru A, Kuscha V, Hans S, Brand M (2017) Ligand-controlled site-specific recombination in zebrafish. In site-specific recombinases (Springer), pp. 87–97
Dai J, Cui X, Zhu Z, Hu W (2010) Non-homologous end joining plays a key role in transgene concatemer formation in transgenic zebrafish embryos. Int J Biol Sci 6:756–768
Felker A, Mosimann C (2016) Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol 135:219–244
Fuchs E, Horsley V (2011) Ferreting out stem cells from their niches. Nat Cell Biol 13:513–518
Hagmann M, Bruggmann R, Xue L, Georgiev O, Schaffner W, Rungger D, Spaniol P, Gerster T (1998) Homologous recombination and DNA-end joining reactions in zygotes and early embryos of zebrafish (Danio rerio) and Drosophila melanogaster. Biol Chem 379:673–681
Hans S, Kaslin J, Freudenreich D, Brand M (2009) Temporally-controlled site-specific recombination in zebrafish. PLoS One 4:e4640
Henninger J, Santoso B, Hans S, Durand E, Moore J, Mosimann C, Brand M, Traver D, Zon L (2017) Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nat Cell Biol 19:17–27
Hoshijima K, Jurynec MJ, Grunwald DJ (2016) Precise editing of the zebrafish genome made simple and efficient. Dev Cell 36:654–667
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Kesavan G, Chekuru A, Machate A, Brand M (2017) CRISPR/Cas9 mediated zebrafish knock-in as a novel strategy to study midbrain-hindbrain boundary development. Front Neuroanat 11:52
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310
Kimura Y, Hisano Y, Kawahara A, Higashijima S (2014) Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4:6545
Kimura Y, Oda M, Nakatani T, Sekita Y, Monfort A, Wutz A, Mochizuki H, Nakano T (2015) CRISPR/Cas9-mediated reporter knock-in in mouse haploid embryonic stem cells. Sci Rep 5:10710
Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S (2011) Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20:713–724
Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M (2011) Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 138:4831–4841
Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258
Li M, Zhao L, Page-McCaw PS, Chen W (2016) Zebrafish genome engineering using the CRISPR–Cas9 system. Trends Genet 32:815–827
Martinez-Barbera JP, Signore M, Boyl PP, Puelles E, Acampora D, Gogoi R, Schubert F, Lumsden A, Simeone A (2001) Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development 128:4789–4800
Ota S, Taimatsu K, Yanagi K, Namiki T, Ohga R, Higashijima SI, Kawahara A (2016) Functional visualization and disruption of targeted genes using CRISPR/Cas9-mediated eGFP reporter integration in zebrafish. Sci Rep 6:34991
Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, Schier AF (2013) Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140:2835–2846
Raible F, Brand M (2004) Divide et Impera—the midbrain-hindbrain boundary and its organizer. Trends Neurosci 27:727–734
Ramachandran, R., Reifler, A., Wan, J., and Goldman, D. (2012). Application of Cre-loxP recombination for lineage tracing of adult zebrafish retinal stem cells. Retinal Development: Methods and Protocols, 884 129–140
Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M (1998) Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125:2381–2395
Rhinn M, Lun K, Amores A, Yan YL, Postlethwait JH, Brand M (2003) Cloning, expression and relationship of zebrafish gbx1 and gbx2 genes to Fgf signaling. Mech Dev 120:919–936
Rhinn M, Picker A, Brand M (2006) Global and local mechanisms of forebrain and midbrain patterning. Curr Opin Neurobiol 16:5–12
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Schier AF, Neuhauss SC, Harvey M, Malicki J, Solnica-Krezel L, Stainier DY, Zwartkruis F, Abdelilah S, Stemple DL, Rangini Z et al (1996) Mutations affecting the development of the embryonic zebrafish brain. Development 123:165–178
Sinha DK, Neveu P, Gagey N, Aujard I, Le Saux T, Rampon C, Gauron C, Kawakami K, Leucht C, Bally-Cuif L (2010) Photoactivation of the CreERT2 recombinase for conditional site-specific recombination with high spatiotemporal resolution. Zebrafish 7:199–204
Sunmonu NA, Li K, Guo Q, Li JY (2011) Gbx2 and Fgf8 are sequentially required for formation of the midbrain-hindbrain compartment boundary. Development 138:725–734
Westerfield, M. (2000) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th edition. University of Oregon Press, Eugene
Zagozewski JL, Zhang Q, Pinto VI, Wigle JT, Eisenstat DD (2014) The role of homeobox genes in retinal development and disease. Dev Biol 393:195–208
Zhou Z, Dang Y, Zhou M, Li L, Yu CH, Fu J, Chen S, Liu Y (2016) Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc Natl Acad Sci U S A 113:E6117–E6125
Acknowledgments
We are thankful to the Chen and Wente labs for providing plasmids to generate Cas9 and sgRNA mRNA (via addgene), Daniela Zoeller for help with cloning the bait plasmid with CreERT2, Dilce Gozuyasli for heat shock experiments, past and present members of the Brand lab for discussions and Vasuprada Iyengar for language and content editing. We thank Marika Fischer, Jitka Michling, Claudia Meyer and Daniela Mögel for dedicated zebrafish care. The Light Microscopy Facility, a core facility of BIOTEC/CRTD at the Technische Universität Dresden, supported this work.
Funding
G.K was supported by post-doctoral fellowships from Swedish research council (Vetenskapsrådet) and an EMBO long-term fellowship (ALTF 350-2011). This work was also supported by an ERC advanced grant (Zf-BrainReg) and project grants of the German Research Foundation (Deutsche Forschungsgemeinschaft, project number BR 1746/6-1 and BR 1746/3) to M.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 225 kb)
Rights and permissions
About this article
Cite this article
Kesavan, G., Hammer, J., Hans, S. et al. Targeted knock-in of CreERT2 in zebrafish using CRISPR/Cas9. Cell Tissue Res 372, 41–50 (2018). https://doi.org/10.1007/s00441-018-2798-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-018-2798-x