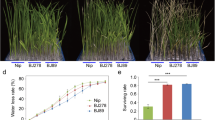
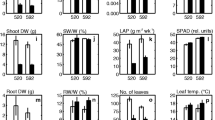
Abstract
Antisense transcription is widespread in many genomes and plays important regulatory roles in gene expression. The objective of our study was to investigate the extent and functional relevance of antisense transcription in forest trees. We employed Populus, a model tree species, to probe the antisense transcriptional response of tree genome under drought, through stranded RNA-seq analysis. We detected nearly 48% of annotated Populus gene loci with antisense transcripts and 44% of them with co-transcription from both DNA strands. Global distribution of reads pattern across annotated gene regions uncovered that antisense transcription was enriched in untranslated regions while sense reads were predominantly mapped in coding exons. We further detected 1185 drought-responsive sense and antisense gene loci and identified a strong positive correlation between the expression of antisense and sense transcripts. Additionally, we assessed the antisense expression in introns and found a strong correlation between intronic expression and exonic expression, confirming antisense transcription of introns contributes to transcriptional activity of Populus genome under drought. Finally, we functionally characterized drought-responsive sense–antisense transcript pairs through gene ontology analysis and discovered that functional groups including transcription factors and histones were concordantly regulated at both sense and antisense transcriptional level. Overall, our study demonstrated the extensive occurrence of antisense transcripts of Populus genes under drought and provided insights into genome structure, regulation pattern and functional significance of drought-responsive antisense genes in forest trees. Datasets generated in this study serve as a foundation for future genetic analysis to improve our understanding of gene regulation by antisense transcription.









Similar content being viewed by others
References
Allen CD, Macalady AK, Chenchouni H, Bachelet D, McDowell N, Vennetier M, Kitzberger T, Rigling A, Breshears DD, Hogg EH Gonzalez P, Fensham R, Zhang Z, Castro J, Demidova N, Lim J-H, Allard G, Running SW, Semerci A, Cobb N (2010) A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest Eco Manag 259:660–684
Amit-Avraham I, Pozner G, Eshar S, Fastman Y, Kolevzon N, Yavin E, Dzikowski R (2015) Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci 112:E982–E991
Anders S, Reyes A, Huber W (2012) Detecting differential usage of exons from RNA-seq data. Genome Res 22:2008–2017
Annilo T, Kepp K, Laan M (2009) Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol 10:81–81
Asensi-Fabado M-A, Amtmann A, Perrella G (2017) Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochim Biophys Acta Gene Regul Mech 1860:106–122
Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu Y-M, Robinson D, Wang R, Chen G, Beer DG et al (2015) The landscape of antisense gene expression in human cancers. Genome Res 25:1068–1079
Beltran M, Puig I, Peña C, García JM, Álvarez AB, Peña R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes Dev 22:756–769
Berta M, Giovannelli A, Sebastiani F, Camussi A, Racchi ML. 2010. Transcriptome changes in the cambial region of poplar (Populus alba L.) in response to water deficit. Plant Biol 12
Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, Guil S (2015) Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci 112:5785–5790
Brosché M, Vinocur B, Alatalo ER, Lamminmaki A, Teichmann T, Ottow EA, Djilianov D, Afif D, Bogeat-Triboulot MB, Altman A (2005) Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol 6:R101
Brunner I, Herzog C, Dawes MA, Arend M, Sperisen C (2015) How tree roots respond to drought. Front Plant Sci 6:547
Carrieri C, Forrest ARR, Santoro C, Persichetti F, Carninci P, Zucchelli S, Gustincich S (2015) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci 9:114
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Chen S, Wang S, Altman A, Hüttermann A (1997) Genotypic variation in drought tolerance of poplar in relation to abscisic acid. Tree Physiol 17:797–803
Chen J, Song Y, Zhang H, Zhang D (2013) Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol Biol Rep 31:946–962
Chen J, Quan M, Zhang D (2015) Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-sEq. Planta 241:125–143
Chen M, Wang C, Bao H, Chen H, Wang Y (2016) Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol Genet Genom 291:1663–1680
Chung PJ, Jung H, Jeong D-H, Ha S-H, Choi YD, Kim J-K (2016) Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom 17:563
Cohen D, Bogeat-Triboulot M-B, Tisserant E, Balzergue S, Martin-Magniette M-L, Lelandais G, Ningre N, Renou J-P, Tamby J-P, Le Thiec D et al (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genom 11:630
Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111:16160–16165
Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10:637–643
Fedak H, Palusinska M, Krzyczmonik K, Brzezniak L, Yatusevich R, Pietras Z, Kaczanowski S, Swiezewski S (2016) Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc Natl Acad Sci 113:E7846–E7855
Gaidatzis D, Burger L, Florescu M, Stadler MB (2015) Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotech 33:722–729
García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A (2012) Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28:2678–2679
Gelfand B, Mead J, Bruning A, Apostolopoulos N, Tadigotla V, Nagaraj V, Sengupta AM, Vershon AK (2011) Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol Cell Biol 31:1701–1709
Guo X-Y, Zhang X-S, Huang Z-Y (2010) Drought tolerance in three hybrid poplar clones submitted to different watering regimes. J Plant Ecol 3:79–87
Hamanishi ET, Raj S, Wilkins O, Thomas BR, Mansfield SD, Plant AL, Campbell MM (2010) Intraspecific variation in the Populus balsamifera drought transcriptome. Plant Cell Environ 33:1742
He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW (2008) The antisense transcriptomes of human cells. Science 322:1855–1857
Hinchee M, Rottmann W, Mullinax L, Zhang C, Chang S, Cunningham M, Pearson L, Nehra N (2009) Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell Dev Biol 45:619–629
Hotta CT, Nishiyama MY Jr, Souza GM (2013) Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS One 8:e71847
Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25:4166–4182
Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J et al (2005) Antisense transcription in the mammalian transcriptome. Science 309:1564–1566
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118
Lembke CG, Nishiyama MY, Sato PM, de Andrade RF, Souza GM (2012) Identification of sense and antisense transcripts regulated by drought in sugarcane. Plant Mol Biol 79:461–477
Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A (2010) Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Meth 7:709–715
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li S, Liberman LM, Mukherjee N, Benfey PN, Ohler U (2013) Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res 23:1730–1739
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Lu T, Zhu C, Lu G, Guo Y, Zhou Y, Zhang Z, Zhao Y, Li W, Lu Y, Tang W et al (2012) Strand-specific RNA-seq reveals widespread occurrence of novel cis- natural antisense transcripts in rice. BMC Genom 13:721
Matsui A, Nguyen AH, Nakaminami K, Seki M (2013) Arabidopsis non-coding RNA regulation in abiotic stress responses. Int J Mol Sci 14:22642–22654
Mills JD, Kawahara Y, Janitz M (2013) Strand-specific RNA-Seq provides greater resolution of transcriptome profiling. Curr Genom 14:173–181
Monclus R, Dreyer E, Villar M, Delmotte FM, Delay D, Petit JM, Barbaroux C, Le Thiec D, Brechet C, Brignolas F (2006) Impact of drought on productivity and water use efficiency in 29 genotypes of Populus deltoides × Populus nigra. New Phytol 169:765
Morrissy AS, Griffith M, Marra MA (2011) Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res 21:1203–1212
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39:877–885
Osato N, Suzuki Y, Ikeo K, Gojobori T (2007) Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 176:1299–1306
Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37:e123–e123
Passalacqua KD, Varadarajan A, Weist C, Ondov BD, Byrd B, Read TD, Bergman NH (2012) Strand-specific RNA-seq reveals ordered patterns of sense and antisense transcription in Bacillus anthracis. PLoS One 7:e43350
Pek JW, Osman I, Tay ML-I, Zheng RT (2015) Stable intronic sequence RNAs have possible regulatory roles in Drosophila melanogaster. J Cell Biol 211:243–251
Pelechano V, Steinmetz LM (2013) Gene regulation by antisense transcription. Nat Rev Genet 14:880–893
Peng C, Ma Z, Lei X, Zhu Q, Chen H, Wang W, Liu S, Li W, Fang X, Zhou X (2011) A drought-induced pervasive increase in tree mortality across Canada's boreal forests. Nat Clim Change 1:467–471
Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM (2007) Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res 35:e128
Phanstiel DH, Boyle AP, Araya CL, Snyder MP, Sushi R (2014) Flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30:2808–2810
Porth I, El-Kassaby YA (2015) Using Populus as a lignocellulosic feedstock for bioethanol. Biotechnol J 10:510–524
Qin T, Zhao H, Cui P, Albesher N, Xiong L (2017) A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. https://doi.org/10.1104/pp.17.00574
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Regier N, Streb S, COCOZZA C, Schaub M, Cherubini P, Zeeman SC, Frey B (2009) Drought tolerance of two black poplar (Populus nigra L.) clones: contribution of carbohydrates and oxidative stress defence. Plant Cell Environ 32:1724–1736
Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AVF, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F et al (2004) Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene 23:6684–6692
Ryan MG (2011) Tree responses to drought. Tree Physiol 31:237–239
Shuai P, Liang D, Zhang Z, Yin W, Xia X (2013) Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom 14:233–233
Shuai P, Liang D, Tang S, Zhang Z, Ye C-Y, Su Y, Xia X, Yin W (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65:4975–4983
Shuai P, Su Y, Liang D, Zhang Z, Xia X, Yin W (2016) Identification of phasiRNAs and their drought- responsiveness in Populus trichocarpa. FEBS Lett 590:3616–3627
Siegel TN, Hon C-C, Zhang Q, Lopez-Rubio J-J, Scheidig-Benatar C, Martins RM, Sismeiro O, Coppée J-Y, Scherf A (2014) Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genom 15:150
Simmons BA, Loque D, Blanch HW (2008) Next-generation biomass feedstocks for biofuel production. Genome Biol 9:242
Street NR, Skogström O, Sjödin A, Tucker J, Rodríguez-Acosta M, Nilsson P, Jansson S, Taylor G (2006) The genetics and genomics of the drought response in Populus. Plant J 48:321–341
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462:799–802
Tian J, Song Y, Du Q, Yang X, Ci D, Chen J, Xie J, Li B, Zhang D (2016) Population genomic analysis of gibberellin-responsive long non-coding RNAs in Populus. J Exp Bot 67:2467–2482
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596
Viger M, Smith HK, Cohen D, Dewoody J, Trewin H, Steenackers M, Bastien C, Taylor G (2016) Adaptive mechanisms and genomic plasticity for drought tolerance identified in European black poplar (Populus nigra L.). Tree Physiol. https://doi.org/10.1093/treephys/tpw017
Wang X-J, Gaasterland T, Chua N-H (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6:R30
Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185
Wang H, Chung PJ, Liu J, Jang I-C, Kean M, Xu J, Chua N-H (2014a) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. https://doi.org/10.1101/gr.165555.113
Wang H, Chung PJ, Liu J, Jang I-C, Kean MJ, Xu J, Chua N-H (2014b) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24:444–453
Wang J, Meng X, Dobrovolskaya OB, Orlov YL, Chen M (2017) Non-coding RNAs and their roles in stress response in plants. Genom Proteom Bioinform 15:301–312
Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM (2009) Genotype and time of day shape the Populus drought response. Plant J 60:703–715
Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875
Xu J, Wang Q, Freeling M, Zhang X, Xu Y, Mao Y, Tang X, Wu F, Lan H, Cao M et al (2017a) Natural antisense transcripts are significantly involved in regulation of drought stress in maize. Nucleic Acids Res 45:5126–5141
Xu W, Xu H, Li K, Fan Y, Liu Y, Yang X, Sun Q (2017b) The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants 3:704–714
Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A (2010) Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11:87
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14–R14
Yuan Y, Chung JD, Fu X, Johnson VE, Ranjan P, Booth SL, Harding SA, Tsai CJ (2009) Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc Natl Acad Sci 106:22020
Zhan S, Lukens L (2013) Protein-coding cis-natural antisense transcripts have high and broad expression in Arabidopsis. Plant Physiol 161:2171–2180
Acknowledgements
We would like to thank Information Technology unit at Michigan Technological University for IT support. We also appreciate prompt technical assistance from the developers and authors of software tools employed in our analysis, including Dr. Douglas H. Phanstiel (Sushi package), Dr. Liguo Wang and Dr. Shengqin Wang (RSeQC), and Dr. Devon Ryan (R script in expression analysis of intronic reads).
Funding
This study is financially supported in part by Michigan Technological University Research Excellence Fund Research Seed Grants and the National Institute of Food and Agriculture, US Department of Agriculture, under Award 2012-67014-19445 to Dr. Yuan.
Author information
Authors and Affiliations
Contributions
YY conceived the study, designed and performed the experiment. YY and SC analyzed the data. YY drafted the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies involving human participants and/or animals.
Dataset access
All sequence reads from this study have been submitted to NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP103219. The raw count data for sense and antisense gene expression of our analysis is available in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE97463. Reviewer can access to the data through https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=etyrokscbhslrop&acc=GSE97463.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, Y., Chen, S. Widespread antisense transcription of Populus genome under drought. Mol Genet Genomics 293, 1017–1033 (2018). https://doi.org/10.1007/s00438-018-1456-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1456-z