Abstract
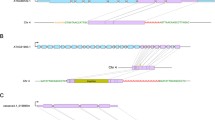
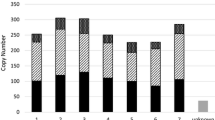
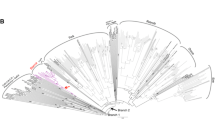
Retrotransposons are ubiquitous components of plant genomes. They affect genome organization, and can also affect the expression patterns of neighboring genes. Retrotransposons are therefore important elements for changing genomic information. To understand the evolution of the Arabidopsis genome, we examined the distribution of certain retrotransposons, AtRE1s and AtRE2s, in the genomes of 12 natural variants (accessions) of Arabidopsis thaliana. AtRE1 and AtRE2 are copia-type retrotransposons that are potentially active. Their copy numbers are low, and they are absent from the genomes of some accessions. We detected four loci with AtRE1s inserted in six accessions, and one locus with an insertion of a solo-LTR-like sequence derived from AtRE1 in two accessions. Seven loci with AtRE2s inserted were detected on eight accessions. These loci were distributed in euchromatic regions of chromosomes 1, 2, 3, and 4. The AtRE1 and AtRE2 sequences at some loci identified in this study have not been recorded in the database of the 1001 Genome project. The sequences of AtRE1s and those of AtRE2s in different accessions and at different loci were highly conserved. There was a complete or almost complete conservation of sequences of both long terminal repeats in each AtRE1 and in each AtRE2. These results suggest that AtRE1 and AtRE2 appeared quite recently in the Arabidopsis genome. Furthermore, sequence comparisons of AtRE1 and AtRE2 loci among accessions revealed the possibility that large deletions containing entire sequences of AtRE1 and AtRE2 have occurred in some accessions.







Similar content being viewed by others
References
Bedbrook JR, Jones J, O’Dell M, Thompson RD, Flavell RB (1980) A molecular description of telometic heterochromatin in Secale species. Cell 19:545–560
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Bucher E, Reinders J, Mirouze M (2012) Epigenetic control of transposons transcription and mobility in Arabidopsis. Curr Opin Plant Biol 15:503–510
Bushman FD (2003) Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135–138
Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Müller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43:956–963
Cheng X, Zhang D, Cheng Z, Keller B, Ling HQ (2009) A new family of Ty1-copia-like retrotransposons originated in the tomato genome by a recent horizontal transfer event. Genetics 181:1183–1193
Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703
Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109:2183–2191
Duan K, Ding X, Zhang Q, Zhu H, Pan A, Huang J (2008) AtCopeg1, the unique gene originated from AtCopia95 retrotransposon family, is sensitive to external hormones and abiotic stresses. Plant Cell Rep 27:1065–1073
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, Kahles A, Bohnert R, Jean G, Derwent P, Kersey P, Belfield EJ, Harberd NP, Kemen E, Toomajian C, Kover PX, Clark RM, Rätsch G, Mott R (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477:419–423
Gao X, Hou Y, Ebina H, Levin HL, Voytas DF (2008) Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res 18:359–369
Girard A, Hannon GJ (2008) Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18:136–148
Gogvadze E, Buzdin A (2009) Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci 66:3727–3742
Grandbastien MA (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3:181–187
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KF, Van de Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481
Huang CRL, Burns KH, Boeke JD (2012) Active transposition in genomes. Annu Rev Genet 46:651–675
Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115–119
Kanazawa A, Liu B, Kong F, Arase S, Abe J (2009) Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J Mol Evol 69:164–175
Kato A, Suzuki M, Kuwahara A, Ooe H, Higano-Inaba K, Komeda Y (1999) Isolation and analysis of cDNA within a 300 kb Arabidopsis thaliana genomic region located around the 100 map unit of chromosome 1. Gene 239:309–316
Kato A, Kato H, Shida T, Saito T, Komeda Y (2009) Evolutionary process of the genomic sequence around the 100 map unit of chromosome 1 in Arabidopsis thaliana. J Plant Biol 52:616–624
Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55:141–172
Kuwahara A, Kato A, Komeda Y (2000) Isolation and characterization of copia-type retrotransposons in Arabidopsis thaliana. Gene 244:127–136
Le QH, Wright S, Yu Z, Bureau T (2000) Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:7376–7381
Lisch D (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60:43–66
Mari-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O (2013) Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45:1029–1039
McCue AD, Slotkin RK (2012) Transposable element small RNAs as regulators of gene expression. Trends Genet 28:616–623
Messing J, Dooner HK (2006) Organization and variability of the maize genome. Curr Opin Plant Biol 9:157–163
Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461:427–430
Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441:947–952
Miura A, Kato M, Watanabe K, Kawabe A, Kotani H, Kakutani T (2004) Genomic localization of endogenous mobile CACTA family transposons in natural variants of Arabidopsis thaliana. Mol Genet Genomics 270:524–532
Pélissier T, Tutois S, Tourmente S, Deragon JM, Picard G (1996) DNA regions flanking the major Arabidopsis thaliana satellite are principally enriched in Athila retroelement sequences. Genetica 97:141–151
Pereira V (2004) Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol 5:R79
Pérez-Hormaeche J, Potet F, Beauclair L, Le Masson I, Courtial B, Bouché N, Lucas H (2008) Invasion of the Arabidopsis genome by the tobacco retrotransposon Tnt1 is controlled by reversible transcriptional gene silencing. Plant Physiol 147:1264–1278
Peterson-Burch BD, Nettleton D, Voytas DF (2004) Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol 5:R78
Reinders J, Wulff B, Mirouze M, Mari-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23:939–950
Sabot F, Schulman AH (2006) Parasitism and the retrotransposon life cycle in plants: a hitchhiker’s guide to the genome. Heredity 97:381–388
Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, Smith LM, Cao J, Fitz J, Warthmann N, Henz SR, Huson DH, Weigel D (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA 108:10249–10254
Suoniemi A, Tanskanen J, Schulman AH (1998) Gypsy-like retrotransposons are widespread in the plant kingdom. Plant J 13:699–705
Takeda S, Sugimoto K, Kakutani T, Hirochika H (2001) Linear DNA intermediates of the Tto1 retrotransposon in Gag particles accumulated in stressed tobacco and Arabidopsis thaliana. Plant J 28:307–317
Terol J, Castillo MC, Bargues M, Pérez-Alonso M, de Frutos R (2001) Structural and evolutionary analysis of the copia-like elements in the Arabidopsis thaliana genome. Mol Biol Evol 18:882–892
Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461:423–426
Tsukahara S, Kawabe A, Kobayashi A, Ito T, Aizu T, Shin-i T, Toyoda A, Fujiyama A, Tarutani Y, Kakutani T (2012) Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev 26:705–713
Vitte C, Panaud O (2005) LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res 110:91–107
Voytas DF, Cummings MP, Koniczny A, Ausubel FM, Rodermel SR (1992) Copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89:7124–7128
Wang W, Zheng H, Fan C, Li J, Shi J, Cai Z, Zhang G, Liu D, Zhang J, Vang S, Lu Z, Wong GK, Long M, Wang J (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18:1791–1802
Wang X, Weigel D, Smith LM (2013) Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet 9:e1003255
Weigel D, Mott R (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10:107
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Wright SI, Agrawal N, Bureau TE (2003) Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res 13:1897–1903
Xiao S, Emerson B, Ratanasut K, Patrick E, O’Neill C, Bancroft I, Turner JG (2004) Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol Biol Evol 21:1662–1672
Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, Navarro L (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110:2389–2394
Zhang X, Wessler SR (2004) Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA 101:5589–5594
Acknowledgments
This work was supported in part by grants-in-aid for scientific research from the Japanese Ministry of Education, Science and Culture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Yamada, M., Yamagishi, Y., Akaoka, M. et al. Genomic localization of AtRE1 and AtRE2, copia-type retrotransposons, in natural variants of Arabidopsis thaliana . Mol Genet Genomics 289, 821–835 (2014). https://doi.org/10.1007/s00438-014-0855-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0855-z