Abstract
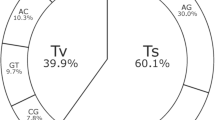
White clover (Trifolium repens L.) is an obligate outbreeding allotetraploid forage legume. Gene-associated SNPs provide the optimum genetic system for improvement of such crop species. An EST resource obtained from multiple cDNA libraries constructed from numerous genotypes of a single cultivar has been used for in silico SNP discovery and validation. A total of 58 from 236 selected sequence clusters (24.5%) were fully validated as containing polymorphic SNPs by genotypic analysis across the parents and progeny of several two-way pseudo-testcross mapping families. The clusters include genes belonging to a broad range of predicted functional categories. Polymorphic SNP-containing ESTs have also been used for comparative genomic analysis by comparison with whole genome data from model legume species, as well as Arabidopsis thaliana. A total of 29 (50%) of the 58 clusters detected putative ortholoci with known chromosomal locations in Medicago truncatula, which is closely related to white clover within the Trifolieae tribe of the Fabaceae. This analysis provides access to translational data from model species. The efficiency of in silico SNP discovery in white clover is limited by paralogous and homoeologous gene duplication effects, which are resolved unambiguously by the transmission test. This approach will also be applicable to other agronomically important cross-pollinating allopolyploid plant species.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 3:525–539
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Attwood SS (1940) Genetics of cross-incompatibility among self-incompatible plants of Trifolium repens. J Am Soc Agron 32:955–968
Barker G, Batley J, O’Sullivan H, Edwards KJ, Edwards D (2003) Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics 19:421–422
Barrett B, Griffiths A, Schreiber M, Ellison N, Mercer C, Bouton J, Ong B, Forster J, Sawbridge T, Spangenberg G, Bryan G, Woodfield D (2004) A microsatellite map of white clover (Trifolium repens L.). Theor Appl Genet 109:596–608
Barrett BA, Baird IJ, Woodfield DR (2005) A QTL analysis of white clover seed production. Crop Sci 5:1844–1850
Batley J, Barker G, O’Sullivan H, Edwards KJ, Edwards D (2003) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 32:84–91
Brennan AC, Harris SA, Hiscock SJ (2006) The population genetics of sporophytic self-incompatibility in Senecio squalidus L. (Asteraceae): S allele diversity across the British range. Evolution 60:213–224
Buetow KH, Edmonson MN, Cassidy AB (1999) Reliable identification of large numbers of candidate SNPs from public EST data. Nat Genet 21:323–325
Burke J, Davison D, Hide W (1999) d2-cluster: a validated method for clustering EST and full-length cDNA sequences. Genome Res 9:1135–1142
Caradus JR, Woodfield DR (1997) World checklist of white clover varieties II. NZ J Agric Res 40:115–206
Chapman R, Erwin TA, Jewell EG, Love CG, Lim GAC, Li X, Mongin M, Spangenberg G, Edwards D (2006) An integrative bioinformatics system for DNA sequence, gene expression, molecular genetic marker, trait and population data. Plant and animal genome XIV, San Diego, p 851
Cogan NOI, Abberton MT, Smith KF, Kearney G, Marshall AH, Williams A, Michaelson-Yeates TPT, Bowen C, Jones ES, Vecchies AC, Forster JW (2006a) Individual and multi-environment combined analyses identify QTLs for morphogenetic and reproductive development traits in white clover (Trifolium repens L.). Theor Appl Genet 112:1401–1415
Cogan NOI, Ponting RC, Vecchies AC, Drayton MC, George J, Dobrowolski MP, Sawbridge TI, Spangenberg GC, Smith KF, Forster JW (2006b) Gene-associated single nucleotide polymorphism (SNP) discovery in perennial ryegrass (Lolium perenne L.). Mol Genet Genomics 276:101–112
Coles ND, Coleman CE, Christensen SA, Jellen EN, Stevens MR, Bonifacio A, Rojas-Beltran JA, Fairbanks DJ, Maughan PJ (2005) Development and use of an expressed sequenced tag library in quinoa (Chenopodium quinoa Willd.) for the discovery of single nucleotide polymorphisms. Plant Sci 168:439–447
Doyle JJ, Luckow MA (2003) The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 131:900–910
Drayton MC, Ponting RC, Vecchies AC, Wilkinson TC, George J, Cogan NOI, Bannan NR, Smith KF, Spangenberg GC, Forster JW (2005) Gene-associated single nucleotide polymorphism discovery in white clover (T. repens L.). In: Humphreys MO (ed) Molecular breeding for the genetic improvement of forage crops and turf. Wageningen Academic Publishers, Netherlands, p 170
Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL (2006) Molecular phylogenetics of the clover genus (Trifolium—Leguminosae). Mol Phylogenet Evol 39:688–705
Forster JW, Jones ES, Kölliker R, Drayton MC, Dupal MP, Guthridge KM, Smith KF, (2001) DNA profiling in outbreeding forage species. In: Henry R (ed) Plant genotyping—the DNA fingerprinting of plants. CABI Press, pp 299–320
Forster JW, Jones ES, Batley J, Smith KF (2004) Molecular marker-based genetic analysis of pasture and turf grasses. In: Hopkins A, Wang Z-Y, Sledge M, Barker RE (eds) Molecular breeding of forage and turf. Kluwer, Dordrecht, pp 197–239
George J, Cogan NOI, Smith KF, Spangenberg GC, Forster JW (2006a) Genetic map integration and comparative genome organisation of white clover (Trifolium repens L.) with model legume species. In: Proceedings of the Third International Legume Genetics and Genomics Conference, Brisbane, April 2006, p 68
George J, Dobrowolski MP, van Zijll de Jong E, Cogan NOI, Smith KF, Forster JW (2006b) Assessment of genetic diversity in cultivars of white clover (Trifolium repens L.) detected by simple sequence repeat polymorphism. Genome 49:919–930
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Guthridge KM, Dupal MD, Kölliker R, Jones ES, Smith KF, Forster JW (2001) AFLP analysis of genetic diversity within and between populations of perennial ryegrass (Lolium perenne L.). Euphytica 122:191–201
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Jones ES, Hughes LJ, Drayton MC, Abberton MT, Michaelson-Yeates TPT, Bowen C, Forster JW (2003) An SSR and AFLP molecular marker-based genetic map of white clover (Trifolium repens L.) Plant Sci 165:447–479
Kölliker R, Jones ES, Drayton MC, Dupal MP, Forster JW (2001) Development and characterisation of simple sequence repeat (SSR) markers for white clover (Trifolium repens L.). Theor Appl Genet 102:416–424
Kota R, Rudd S, Facius A, Kolesov G, Thiel T, Zhang H, Stein N, Mayer K, Graner A (2003) Snipping polymorphisms from large EST collections in barley (Hordeum vulgare L.). Mol Gen Genomics 270:24–33
Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook DR, Huguet T, de Jong JH, Fransz PF, Bisseling T (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27:49–58
Labate JA, Baldo AM (2005) Tomato SNP discovery by EST mining and resequencing. Mol Breed 16:343–349
Lopez C, Piégu B, Cooke R, Delseny M, Tohme J, Verdier V (2005) Using cDNA and genomic sequences as tools to develop SNP strategies in cassava (Manihot esculenta Crantz). Theor Appl Genet 110:425–431
Love CG, Batley J, Edwards D (2003) Applied computational tools for crop genome research. J Plant Biotech 5:193–105
Morales M, Roig E, Monforte AJ, Arús P, Garcia-Mas J (2004) Single-nucleotide polymorphisms detected in expressed sequence tags of melon (Cucumis melo L.). Genome 47:352–360
Ogihara Y, Hasegawa K, Tsuijimoto H (1994) High-resolution cytological mapping of the long arm of chromosome 5A in common wheat using a series of deletion lines induced by gametocidal (Gc) genes of Aegilops speltoides. Mol Gen Genomics 244:253–259
Pérez-Collazos E, Catalán P (2006) Palaeopolyploidy, spatial structure and conservation genetics of the narrow steppe plant Vella pseudocytisus subsp. paui (Vellinae, Cruciferae). Ann Bot 97:635–647
Picoult-Newberg L, Ideker TE, Pohl MG, Taylor SL, Donaldson MA, Nickerson DA, Boyce-Jacino M (1999) Mining SNPs from EST databases. Genome Res 9:167–176
Ritter E, Gebhardt C, Salamini F (1990) Estimation of recombination frequencies and construction of linkage maps from crosses between heterozygous parents. Genetics 125:645–654
Rogers ME, Noble CL, Halloran GM, Nicolas ME (1997) Selecting for salt tolerance in white clover (Trifolium repens): chloride ion exclusion and its heritability. New Phytol 135:645–654
Rostocks N, Mudie S, Cardle L, Russell J, Ramsay L, Booth A, Svensson JT, Wanamaker SI, Walia H, Rodriguez EM, Hedley PE, Liu H, Morris J, Close TJ, Marshall DF, Waugh R (2005) Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Mol Gen Genomics 274:515–527
Sawbridge T, Ong E-K, Binnion C, Emmerling M, Meath K, Nunan K, O’Neill M, O’Toole F, Simmonds J, Wearne K, Winkworth A, Spangenberg G (2003a) Generation and analysis of expressed sequence tags in white clover (Trifolium repens L.)Plant Sci 165:1077–1087
Sawbridge T, Ong E-K, Binnion C, Emmerling M, McInnes R, Meath K, Nguyen N, Nunan K, O’Neill M, O’Toole F, Rhodes C, Simmonds J, Tian P, Wearne K, Webster T, Winkworth A, Spangenberg G (2003b) Generation and analysis of expressed sequence tags in perennial ryegrass (Lolium perenne L.). Plant Sci 165:1089–1100
Schmid KJ, Sörensen TR, Stracke R, Törjerk O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13:1250–1257
Somers DJ, Kirkpatrick R, Moniwa M, Walsh A (2003) Mining single-nucleotide polymorphisms from hexaploid wheat ESTs. Genome 49:431–437
Sorrells ME, Wilson WA (1997) Direct classification and selection of superior alleles for crop improvement. Crop Sci 37:691–697
Spangenberg G, Forster JW, Edwards D, John U, Mouradov A, Emmerling M, Batley J, Felitti S, Cogan NOI, Smith KF, Dobrowolksi MP (2005a) Future directions in the molecular breeding of forage and turf. In: Humphreys MO (ed) Molecular breeding for the genetic improvement of forage crops and turf. Wageningen Academic Publishers, Netherlands, pp 83–97
Spangenberg G, Sawbridge T, Ong EK, Love CG, Erwin TA, Logan EG, Edwards D (2005b) Clover ASTRA: a web-based resource for Trifolium EST analysis. In: Humphreys MO (ed) Molecular breeding for the genetic improvement of forage crops and turf. Wageningen Academic Publishers, Netherlands, p 195
Spangenberg G, Sawbridge T, Ong EK, Love CG, Erwin TA, Logan EG, Edwards D (2005c) Ryegrass ASTRA: a web-based resource for Lolium EST analysis. In: Humphreys MO (ed) Molecular breeding for the genetic improvement of forage crops and turf. Wageningen Academic Publishers, Netherlands, p 201
Taillon-Miller P, Gu Z, Li Q, Hillier L, Kwok P-Y (1998) Overlapping genomic sequences: a treasure trove of single-nucleotide polymorphisms. Genome Res 8:748–754
Williams W (1987) Adaptive variation. In: Baker MJ, Williams WM (eds) White clover. CAC International, Wallingford, pp 299–321
Yang WC, Bai X-D, Kabelka E, Eaton C, Kamoun S, van der Knapp E, Francis D (2004) Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Mol Breed 14:21–34
Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S. (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137:1174–1181
Zhu H, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organisation of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Micro Interact 15:529–539
Zhu H, Choi H-K, Cook DR, Shoemaker RC (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137:1189–1196
Acknowledgments
This work was supported by funding from the Victorian Department of Primary Industries, Dairy Australia Ltd., the Geoffrey Gardiner Dairy Foundation, Meat and Livestock Australia Ltd. and the Molecular Plant Breeding Cooperative Research Centre. The authors thank Prof. Michael Hayward for careful critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W.R. McCombie.
N.O.I. Cogan and M.C. Drayton contributed equally to this work.
Electronic supplementary material
Below are the links to the electronic supplementary materials.
Rights and permissions
About this article
Cite this article
Cogan, N.O.I., Drayton, M.C., Ponting, R.C. et al. Validation of in silico-predicted genic SNPs in white clover (Trifolium repens L.), an outbreeding allopolyploid species. Mol Genet Genomics 277, 413–425 (2007). https://doi.org/10.1007/s00438-006-0198-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0198-5