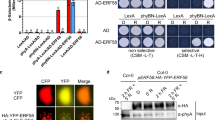
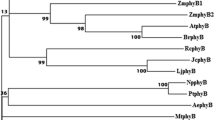
Abstract
Phytochrome photoreceptors enable plants to perceive divergent light signals leading to adaptive changes in response to differing environmental conditions. However, the mechanism of light signal transduction is not fully understood. Here we report the identification of a new signaling intermediate from Arabidopsis thaliana, Scarecrow-like (SCL)13, which serves as a positive regulator of continuous red light signals downstream of phytochrome B (phyB). SCL13 antisense lines exhibit reduced sensitivity towards red light, but only a distinct subset of phyB-mediated responses is affected, indicating that SCL13 executes its major role in hypocotyl elongation during de-etiolation. Genetic evidence suggests that SCL13 is also needed to modulate phytochrome A (phyA) signal transduction in a phyB-independent way. The SCL13 protein is localized in the cytoplasm, but can also be detected in the nucleus. Overexpression of both a nuclear and cytoplasmic localized SCL13 protein leads to a hypersensitive phenotype under red light indicating that SCL13 is biologically active in both compartments. SCL13 is a member of the plant-specific GRAS protein family, which is involved in various different developmental and signaling pathways. A previously identified phytochrome A signaling intermediate, PAT1, belongs to the same subbranch of GRAS proteins as SCL13. Although both proteins are involved in phytochrome signaling, each is specific for a different light condition and regulates a different subset of responses.








Similar content being viewed by others
References
Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9:1317–1326
Benditt JO, Meyer C, Fasold H, Barnard FC, Riedel N (1989) Interaction of a nuclear location signal with isolated nuclear envelopes and identification of signal-binding proteins by photoaffinity labeling. Proc Natl Acad Sci USA 86:9327–9331
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14:1269–1278
Canton FR, Quail PH (1999) Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol 121:1207–1216
Casal JJ (1996) Phytochrome A enhances the promotion of hypocotyl growth caused by reductions in levels of phytochrome B in its far-red-light-absorbing form in light-grown Arabidopsis thaliana. Plant Physiol 112:965–973
Casal JJ, Yanovsky MJ, Luppi JP (2000) Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem Photobiol 71:481–486
Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423:881–885
Cerdan PD, Yanovsky MJ, Reymundo FC, Nagatani A, Staneloni RJ, Whitelam GC, Casal JJ (1999) Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J 18:499–507
Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38:87–117
Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401:610–613
Choi G, Kim JI, Hong SW, Shin B, Choi G, Blakeslee JJ, Murphy AS, Seo Y, Kim K, Koh EJ, Song PS, Lee H (2005) A possible role of NDPK2 in the regulation of auxin mediated responses for plant growth and development. Plant Cell Physiol 46:1246–1254
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25:413–427
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Devlin PF, Halliday KJ, Harberd NP, Whitelam GC (1996) The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: novel phytochromes control internode elongation and flowering time. Plant J 10:1127–1134
Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10:1479–1487
Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119:909–915
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433
Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419:74–77
Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10:51–54
Fankhauser C (2002) Light perception in plants: cytokinins and red light join forces to keep phytochrome B active. Trends Plant Sci 7:143–45
Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216:1–16
Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284:1539–1541
Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18:4679–4688
Franklin KA, Whitelam GC (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot (Lond) 96:169–175
Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003a) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15:1981–1989
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003b) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131:1340–1346
Furuya M, Schäfer E (1996) Photoperception and signalling of induction reactions by different phytochromes. Trends Plant Sci 1:301–307
Goda H, Shimada Y, Fujioka S, Yoshida S (2004) Classification of brassinosteroid-regulated genes based on expression profiles in bri1 and in response to a protein kinase inhibitor, staurosporin. Biosci Biotechnol Biochem 68:1605–1607
Greb T, Clarenz O, Schäfer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17:1175–1187
Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99:11513–11518
Gyula P, Schäfer E, Nagy F (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6:446–452
Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131:1913–1920
Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101:555–567
Hennig L, Poppe C, Unger S, Schäfer E (1999) Control of hypocotyl elongation in Arabidopsis thaliana by photoreceptor interaction. Planta 208:257–263
Hennig L, Poppe C, Sweere U, Martin A, Schäfer E (2001) Negative interference of endogenous phytochrome B with phytochrome A function in Arabidopsis. Plant Physiol 125:1036–1044
Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schäfer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128:194–200
Hoecker U, Toledo-Ortiz G, Bender J, Quail PH (2004) The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219:195–200
Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16:1247–1259
Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97:9789–9794
Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Khanna R, Kikis EA, Quail PH (2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133:1530–1538
Kim SH, Roux SJ (2003) An Arabidopsis Ran-binding protein, AtRanBP1c, is a co-activator of Ran GTPase-activating protein and requires the C-terminus for its cytoplasmic localization. Planta 216:1047–1052
Kim DH, Kang JG, Yang SS, Chung KS, Song PS, Park CM (2002) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14:3043–3056
Kircher S, Gil P, Kozma-Bognar L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adam E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14:1541–1555
Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16:393–401
van der Krol AR, Chua NH (1991) The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell 3:667–675
Lim J, Jung JW, Lim CE, Lee MH, Kim BJ, Kim M, Bruce WB, Benfey PN (2005) Conservation and diversification of SCARECROW in maize. Plant Mol Biol. 59:619–630
Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13:1293–1304
Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15:223–236
Millar AJ (2003) A suite of photoreceptors entrains the plant circadian clock. J Biol Rhythms 18:217–226
Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15:1962–1980
Morelli G, Ruberti I (2002) Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci 7:399–404
Nagatani A (2004) Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol 7:708–711
Nagy F, Schäfer E (2000) Nuclear and cytosolic events of light-induced, phytochrome-regulated signaling in higher plants. EMBO J 19:157–163
Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53:329–355
Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005) The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol 46:609–619
Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118:27–35
Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95:657–667
Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11:2983–2995
Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG (2003) The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34:161–171
Patschinsky T, Hunter T, Esch FS, Cooper JA, Sefton BM (1982) Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc Natl Acad Sci USA 79:973–977
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18:111–119
Quail PH (1997) The phytochromes: a biochemical mechanism of signaling in sight? Bioessays 19:571–579
Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3:85–93
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147–157
Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, Millar AJ (2000) Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol 122:1149–1160
Robson P, Whitelam GC, Smith H (1993) Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in Phytochrome B. Plant Physiol 102:1179–1184
Sakamoto K, Nagatani A (1996) Nuclear localization activity of phytochrome B. Plant J 10:859–868
Schäfer E, Bowler C (2002) Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep 3:1042–1048
Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93:1219–1229
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37:501–506
Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96:290–295
Schwechheimer C, Schwager K (2004) Regulated proteolysis and plant development. Plant Cell Rep 23:353–364
Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130:442–456
Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93:8129–8133
Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155–169
Smith H (1999) Phytochromes. Tripping the light fantastic. Nature 400:710–711, 713
Smith H (2000) Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407:585–591
Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20:840–844
Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101:319–329
Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C (2003) The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev 17:256–268
Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126:4235–4245
Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289:768–771
Stuurman J, Jaggi F, Kuhlemeier C (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16:2213–2218
Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260:289–297
Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schäfer E, Harter K (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294:1108–1111
Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98:9437–9442
Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38:725–739
Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14:301–319
Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54:519–532
Wagner D, Koloszvari M, Quail PH (1996) Two Small Spatially Distinct Regions of Phytochrome B Are Required for Efficient Signaling Rates. Plant Cell 8:859–871
Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9:491–507
Wang H, Ma L, Habashi J, Li J, Zhao H, Deng XW (2002) Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32:723–733
Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof Transcription Factor OBP3 Modulates Phytochrome and Cryptochrome Signaling in Arabidopsis. Plant Cell 17:475–485
Zhu Y, Tepperman JM, Fairchild CD, Quail PH (2000) Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci USA 97:13419–13424
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgments
We would like to thank Prof. Dr. R. G Herrmann for his support, Christine Matzenbacher for excellent technical assistance, and Randy Foster for helpful discussions and suggestions. This work was supported by a grant from the “Deutsche Forschungsgemeinschaft” (DFG) to C.B. (Bo1146/3). Work at Rockefeller was supported by NIH grant GM 44640 to N.H.C.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Jürgens
Rights and permissions
About this article
Cite this article
Torres-Galea, P., Huang, LF., Chua, NH. et al. The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol Genet Genomics 276, 13–30 (2006). https://doi.org/10.1007/s00438-006-0123-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0123-y