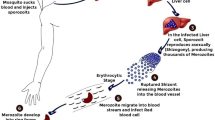
Abstract
The apicomplexan parasite Plasmodium falciparum is responsible for global malaria burden. With the reported resistance to artemisinin chemotherapy, there is an urgent need to maintain early phase drug discovery and identify novel drug targets for successful eradication of the pathogen from the host. In our previous work on comparative genomics study for identification of putative essential genes and therapeutic candidates in P. falciparum, we predicted 11 proteins as anti-malarial drug targets from PlasmoDB database. In this paper, we made an attempt for identification of novel drug targets in P. falciparum genome using a sequence of computational methods from Malaria Parasite Metabolic Pathway database. The study reported the identification of 71 proteins as potential drug targets for anti-malarial interventions. Furthermore, homology modeling and molecular dynamic simulation study of one of the potential drug targets, aminodeoxychorismate lyase, was carried to predict the 3D structure of the protein. Structure and ligand-based drug designing reported MMV019742 from Pathogen Box and TCAMS-141515 from GSK-TCAMS library as potential hits. The reliability of the binding mode of the inhibitors is confirmed by GROMACS for a simulation time of 20 ns in water environment. This will be helpful for experimental validation of the small-molecule inhibitor.







Similar content being viewed by others
References
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Lim P (2014) A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55
Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Heiges M (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res 37:D539–D543
Barmade MA, Murumkar PR, Kumar Sharma M, Shingala KP, Giridhar RR, Yadav MR (2015) Discovery of anti-malarial agents through application of in silico studies. Comb Chem High Throughput Screen 18:151–187
Berendsen HJ, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Tasumi M (1978) The Protein Data Bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys 185:584–559
Bhasin M, Raghava GPS (2004) ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res 32:W414–W419
Camach C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10(1):421
Chang TH, Wu LC, Lee TY, Chen SP, Huang HD, Horng JT (2013) EuLoc: a web-server for accurately predict protein subcellular localization in eukaryotes by incorporating various features of sequence segments into the general form of Chou’s PseAAC. J Comput Aided Mol Des 27(1):91–103
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an n.Log (N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Elliott DA, McIntosh MT, Hosgood HD, Chen S, Zhang G, Baevova P, Joiner KA (2008) Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum. PNAS 105(7):2463–2468
Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Brown JR (2010) Thousands of chemical starting points for antimalarial lead identification. Nature 465(7296):305–310
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11(6):681–684
Ginsburg H (2006) Progress in in silico functional genomics: the malaria metabolic pathways database. Trends Parasitol 22(6):238–240
Ginsburg H (2009) Caveat emptor: limitations of the automated reconstruction of metabolic pathways in Plasmodium. Trends Parasitol 25(1):37–43
Ginsburg H, Tilley L (2011) Plasmodium falciparum metabolic pathways (MPMP) project upgraded with a database of subcellular locations of gene products. Trends Parasitol 27(7):285–286
Hess B, Bekker H, Berendsen HJ, Fraaije JG (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Finn RD (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215
Hyde JE (2005) Exploring the folate pathway in Plasmodium falciparum. Acta Trop 94(3):191–206
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Jorgensen WL, Tirado-Rives J (1988) The OPLS [optimized potentials for liquid simulations] potential functons for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110(6):1657–1666
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereo chemical quality of protein structures. J App Crystallogr 26(2):283–291
Lemkul JA, Allen WJ, Bevan DR (2010) Practical considerations for building GROMOS-compatible small-molecule topologies. J Chem Informat Mod 50(12):2221–2235
Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13(9):2178–2189
Ludin P, Woodcroft B, Ralph SA, Mäser P (2012) In silico prediction of antimalarial drug target candidates. International Journal for Parasitology: Drugs and Drug Resistance 2:191–199
Lu F, Jiang H, Ding J, Mu J, Valenzuela JG, Ribeiro JM, Su XZ (2007) cDNA sequences reveal considerable gene prediction inaccuracy in the Plasmodium falciparum genome. BMC Genomics 8(1):255
Magnani G, Lomazzi M, Peracchi A (2013) Completing the folate biosynthesis pathway in Plasmodium falciparum: p-aminobenzoate is produced by a highly divergent promiscuous aminodeoxychorismate lyase. Biochem J 455(2):149–155
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55(14):6582–6594
O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) OpenBabel: an open chemical toolbox. Journal of Chemoinformatics 3(1):33
Percudani R, Peracchi A (2003) A genomic overview of pyridoxal phosphate dependent enzymes. EMBO Rep 4(9):850–854
Pierleoni A, Martelli PL, Fariselli P, Casadio R (2006) BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22(14):e408–e416
Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Webb B (2004) MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res 32:D217–D222
Rout S, Warhurst DC, Suar M, Mahapatra RK (2015) In-silico comparative genomics analysis of Plasmodium falciparum for the identification of putative essential genes and therapeutic candidates. J Microbio Meth 109:1–8
Rout S, Mahapatra RK (2016) In silico screening of novel inhibitors of M17 leucine amino peptidase (LAP) of Plasmodium vivax as therapeutic candidate. Biomed & Pharmacoth 82:192–201
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5(4):725–738
Sali A, Blundell T (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Bio 5:779–815
SchuÈttelkopf AW, Van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallograph 60(8):1355–1363
Shanmugham B, Pan A (2013) Identification and characterization of potential therapeutic candidates in emerging human pathogen Mycobacterium abscessus: a novel hierarchical in silico approach. PLoS One 8(3):e59126
Spangenberg T, Burrows JN, Kowalczyk P, McDonald S, Wells TN, Willis P (2013) The open access malaria box: a drug discovery catalyst for neglected diseases. PLoS One 8(6):e62906
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Jensen LJ (2011) The STRING database in 2011: functional interaction networks of proteins globally integrated and scored. Nucleic Acids Res 39:D561–D568
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein–ligand docking using GOLD. Proteins 52(4):609–623
Verdonk ML, Berdini V, Hartshorn MJ, Mooij WT, Murray CW, Taylor RD, Watson P (2004) Virtual screening using protein−ligand docking: avoiding artificial enrichment. J Chem Inf Comp Sci 44(3):793–806
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718
Vaught A (1996) Graphing with Gnuplot and Xmgr: two graphing packages available under linux. Linux J 1996(28es):7
Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res 38:W469–W473
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45(1):160–169
WHO (2015) World malaria report. World Health Organization, Geneva
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534
Yuthavong Y, Kamchonwongpaisan S, Leartsakulpanich U, Chitnumsub P (2006) Folate metabolism as a source of molecular targets for antimalarials. Future Microbiol 1(1):113–125
Acknowledgements
The authors would like to acknowledge the Bioinformatics Lab Facility of School of Biotechnology, KIIT University, during the course of the work. Miss Subhashree Rout would like to acknowledge DST, Government of India, for the financial support to pursue her PhD work through INSPIRE fellowship. SR and RKM thank Prof. Mrutyunjay Suar, Director of School of Biotechnology in KIIT University, for his encouragement and support during the course of study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Table 1
(DOCX 11 kb)
Supplementary Table 2
(DOCX 11 kb)
Supplementary Table 3
(DOCX 11 kb)
Supplementary Table 4
(DOCX 12 kb)
Supplementary Fig. 1
(DOCX 151 kb)
Supplementary Fig. 2
(DOCX 46 kb)
Supplementary Fig. 3
(DOCX 719 kb)
Supplementary Fig. 4
(DOCX 452 kb)
Supplementary Fig. 5
(DOCX 176 kb)
Rights and permissions
About this article
Cite this article
Rout, S., Patra, N.P. & Mahapatra, R.K. An in silico strategy for identification of novel drug targets against Plasmodium falciparum . Parasitol Res 116, 2539–2559 (2017). https://doi.org/10.1007/s00436-017-5563-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5563-2