Abstract
Objectives
Increasing evidence suggests that long non-coding RNAs (lncRNAs) may play a crucial role in many biological processes in a variety of cancers and serve as the basis for many clinical applications including prognostic biomarkers and potential therapeutic targets. The aim of this study is to develop a prognostic lncRNA signature with RNA-seq data in lung adenocarcinomas.
Methods
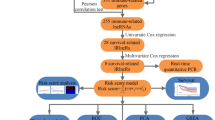
LncRNA expression profiles and clinical data of lung adenocarcinoma patients from The Cancer Genome Atlas (TCGA) were analyzed. Univariate Cox proportional regression model was used to identify prognostic lncRNAs, and then multivariate Cox proportional regression model was used to develop a prognostic signature. Survivals were compared using log-rank test, and the biological implications of prognostic lncRNAs were analyzed using the KEGG pathway functional enrichment analysis.
Results
We identified eight lncRNAs which had prognostic association with p value <0.01 in a TCGA lung adenocarcinoma cohort of 478 patients. Then a novel prognostic signature with the eight lncRNAs was developed using Cox regression model. Signature high-risk cases had worse overall survival (OS, median 85.97 vs. 38.34 months, p < 0.001) and disease-free survival (DFS, median 44.02 vs. 26.58 months, p = 0.007) than low-risk cases. Multivariate Cox regression analysis suggested that the eight-lncRNA signature was independent of clinical and pathological factors. KEGG pathway functional enrichment analysis indicated potential functional roles of the eight prognostic lncRNAs in tumorigenesis.
Conclusions
Our findings suggest that the eight-lncRNA signature might provide an effective independent prognostic model for the prediction of lung adenocarcinoma patients.






Similar content being viewed by others
References
Beer DG, Kardia SLR, Huang C, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JMG, Iannettoni MD, Orringer MB, Hanash S (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8(8):816–824. doi:10.1038/nm733
Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga J, Brain E, Causeret S, DeLorenzi M, Glas AM, Golfinopoulos V, Goulioti T, Knox S, Matos E, Meulemans B, Neijenhuis PA, Nitz U, Passalacqua R, Ravdin P, Rubio IT, Saghatchian M, Smilde TJ, Sotiriou C, Stork L, Straehle C, Thomas G, Thompson AM, van der Hoeven JM, Vuylsteke P, Bernards R, Tryfonidis K, Rutgers E, Piccart M (2016) 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med 375(8):717–729. doi:10.1056/NEJMoa1602253
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
Chandra GS, Nandan TY (2016) Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. doi:10.1002/ijc.30546
Chen H, Yu S, Chen C, Chang G, Chen C, Yuan A, Cheng C, Wang C, Terng H, Kao S, Chan W, Li H, Liu C, Singh S, Chen WJ, Chen JJW, Yang P (2007a) A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356(1):11–20. doi:10.1056/NEJMoa060096
Chen H, Yu S, Chen C, Chang G, Chen C, Yuan A, Cheng C, Wang C, Terng H, Kao S, Chan W, Li H, Liu C, Singh S, Chen WJ, Chen JJW, Yang P (2007b) A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356(1):11–20. doi:10.1056/NEJMoa060096
Endoh H (2004) Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol 22(5):811–819. doi:10.1200/JCO.2004.04.109
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):l1. doi:10.1126/scisignal.2004088
Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21(11):1253–1261. doi:10.1038/nm.3981
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu Y, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47(3):199–208. doi:10.1038/ng.3192
Kaczkowski B, Tanaka Y, Kawaji H, Sandelin A, Andersson R, Itoh M, Lassmann T, Hayashizaki Y, Carninci P, Forrest ARR (2016) Transcriptome analysis of recurrently deregulated genes across multiple cancers identifies new pan-cancer biomarkers. Can Res 76(2):216–226. doi:10.1158/0008-5472.CAN-15-0484
Kawaguchi A, Iwadate Y, Komohara Y, Sano M, Kajiwara K, Yajima N, Tsuchiya N, Homma J, Aoki H, Kobayashi T, Sakai Y, Hondoh H, Fujii Y, Kakuma T, Yamanaka R (2012) Gene expression signature-based prognostic risk score in patients with primary central nervous system lymphoma. Clin Cancer Res 18(20):5672–5681. doi:10.1158/1078-0432.CCR-12-0596
Kratz JR, Mann MJ, Jablons DM (2013) International trial of adjuvant therapy in high risk stage I non-squamous cell carcinoma identified by a 14-gene prognostic signature. Transl Lung Cancer Res 2(3):222–225. doi:10.3978/j.issn.2218-6751.2013.05.01
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669. doi:10.1534/genetics.112.146704
Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Can Res 75(18):3728–3737. doi:10.1158/0008-5472.CAN-15-0273
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW (2016) Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci 20(11):2285–2295
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A (2016) Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66(4):271–289. doi:10.3322/caac.21349
Niedzwiecki D, Frankel WL, Venook AP, Ye X, Friedman PN, Goldberg RM, Mayer RJ, Colacchio TA, Mulligan JM, Davison TS, O’Brien E, Kerr P, Johnston PG, Kennedy RD, Harkin DP, Schilsky RL, Bertagnolli MM, Warren RS, Innocenti F (2016) Association between results of a gene expression signature assay and recurrence-free interval in patients with stage II colon cancer in cancer and Leukemia Group B 9581 (alliance). J Clin Oncol 34(25):3047–3053. doi:10.1200/JCO.2015.65.4699
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29(4):452–463. doi:10.1016/j.ccell.2016.03.010
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102(43):15545–15550. doi:10.1073/pnas.0506580102
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108. doi:10.3322/caac.21262
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I (2015) The 2015 world health organization classification of lung tumors. J Thorac Oncol 10(9):1243–1260. doi:10.1097/JTO.0000000000000630
Wahlestedt C (2013) Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discovery 12(6):433–446. doi:10.1038/nrd4018
Wang L, He Y, Liu W, Bai S, Xiao L, Zhang J, Dhanasekaran SM, Wang Z, Kalyana-Sundaram S, Balbin OA, Shukla S, Lu Y, Lin J, Reddy RM, Carrott PJ, Lynch WR, Chang AC, Chinnaiyan AM, Beer DG, Zhang J, Chen G (2016) Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 7(10):11487–11499. doi:10.18632/oncotarget.7203
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan C, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L (2015) Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28(4):529–540. doi:10.1016/j.ccell.2015.09.006
Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J (2015) A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med 13(1):231. doi:10.1186/s12967-015-0556-3
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, S., Zheng, D., Dong, C. et al. Development of a novel prognostic signature of long non-coding RNAs in lung adenocarcinoma. J Cancer Res Clin Oncol 143, 1649–1657 (2017). https://doi.org/10.1007/s00432-017-2411-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-017-2411-9