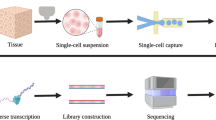
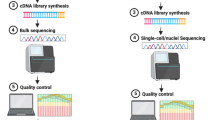
Abstract
Exploring and revealing the secret of the function of the human brain has been the dream of mankind and science. Delineating brain transcriptional regulation has been extremely challenging, but recent technological advances have facilitated a deeper investigation of molecular processes in the brain. Tracing the molecular regulatory mechanisms of different gene expression profiles in the brain is divergent and has made it possible to connect spatial and temporal variations in gene expression to distributed properties of brain structure and function. Here, we review the molecular diversity of the brain among rodents, non-human primates and humans. We also discuss the molecular mechanism of non-coding DNA/RNA at the transcriptional/post-transcriptional level based on recent technical advances to highlight an improved understanding of the complex transcriptional network in the brain. Spatiotemporal and single-cell transcriptomics have attempted to gain novel insight into the development and evolution of the brain as well as the progression of human diseases. Although it is clear that the field is developing and challenges remain to be resolved, the impressive recent progress provides a solid foundation to better understand the brain and evidence-based recommendations for the diagnosis and treatment of brain diseases.


Similar content being viewed by others
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005
Amunts K, Zilles K (2015) Architectonic mapping of the human brain beyond Brodmann. Neuron 88:1086–1107
Amunts K, Mohlberg H, Bludau S, Zilles K (2020) Julich-Brain: a 3D probabilistic atlas of the human brain’s cytoarchitecture. Science 369:988–992
Arnold N, Girke T, Sureshchandra S, Messaoudi I (2016) Acute Simian varicella virus infection causes robust and sustained changes in gene expression in the sensory Ganglia. J Virol 90:10823–10843
Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M et al (2016) Evolution of osteocrin as an activity-regulated factor in the primate brain. Nature 539:242–247
Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA et al (2016) A comprehensive transcriptional map of primate brain development. Nature 535:367–375
Barouch DH, Ghneim K, Bosche WJ, Li Y, Berkemeier B et al (2016) Rapid inflammasome activation following mucosal SIV infection of rhesus monkeys. Cell 165:656–667
Barter J, Kumar A, Stortz JA, Hollen M, Nacionales D et al (2019) Age and sex influence the hippocampal response and recovery following sepsis. Mol Neurobiol 56:8557–8572
Bludau S, Muhleisen TW, Eickhoff SB, Hawrylycz MJ, Cichon S et al (2018) Integration of transcriptomic and cytoarchitectonic data implicates a role for MAOA and TAC1 in the limbic-cortical network. Brain Struct Funct 223:2335–2342
Boddy AM, Harrison PW, Montgomery SH, Caravas JA, Raghanti MA et al (2017) Evidence of a conserved molecular response to selection for increased brain size in primates. Genome Biol Evol 9:700–713
Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD et al (2018) Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci 21:1185–1195
Bose DA, Berger SL (2017) eRNA binding produces tailored CBP activity profiles to regulate gene expression. RNA Biol 14:1655–1659
Breschi A, Gingeras TR, Guigo R (2017) Comparative transcriptomics in human and mouse. Nat Rev Genet 18:425–440
Carter RA, Bihannic L, Rosencrance C, Hadley JL, Tong Y et al (2018) A single-cell transcriptional atlas of the developing murine cerebellum. Curr Biol 28:2910-2920.e2
Chen W, Qin C (2015) General hallmarks of microRNAs in brain evolution and development. RNA Biol 12:701–708
Chen J, Suo S, Tam PP, Han JJ, Peng G et al (2017) Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat Protoc 12:566–580
Chen F, Zhang L, Wang E, Zhang C, Li X (2018a) LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem Biophys Res Commun 496:184–190
Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB et al (2018b) Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet 50:1296–1303
Chiu YC, Wang LJ, Lu TP, Hsiao TH, Chuang EY et al (2017) Differential correlation analysis of glioblastoma reveals immune ceRNA interactions predictive of patient survival. BMC Bioinform 18:132
Cui B, Li B, Liu Q, Cui Y (2017) lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem 118:4548–4557
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T et al (2012) Landscape of transcription in human cells. Nature 489:101–108
Dweep H, Gretz N, Sticht C (2014) miRWalk database for miRNA-target interactions. Methods Mol Biol 1182:289–305
Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK et al (2016) Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19:504–516
Hanan M, Soreq H, Kadener S (2017) CircRNAs in the brain. RNA Biol 14:1028–1034
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L et al (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489:391–399
Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T et al (2015) Canonical genetic signatures of the adult human brain. Nat Neurosci 18:1832–1844
He Z, Han D, Efimova O, Guijarro P, Yu Q et al (2017) Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat Neurosci 20:886–895
Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER et al (2019) Conserved cell types with divergent features in human versus mouse cortex. Nature 573:61–68
Huisman SMH, van Lew B, Mahfouz A, Pezzotti N, Hollt T et al (2017) BrainScope: interactive visual exploration of the spatial and temporal human brain transcriptome. Nucleic Acids Res 45:e83
Jager J, Marwitz S, Tiefenau J, Rasch J, Shevchuk O et al (2014) Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun 82:275–285
Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC et al (2012) Human-specific transcriptional networks in the brain. Neuron 75:601–617
Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC (2016) Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci USA 113:E469-478
Labonte B, Engmann O, Purushothaman I, Menard C, Wang J et al (2017) Sex-specific transcriptional signatures in human depression. Nat Med 23:1102–1111
Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC et al (2016) Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352:1586–1590
Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A et al (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176
Lein E, Borm LE, Linnarsson S (2017) The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358:64–69
Li J, Tan S, Kooger R, Zhang C, Zhang Y (2014a) MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev 43:506–517
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014b) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92-97
Lin D, Liang Y, Jing X, Chen Y, Lei M et al (2018) Microarray analysis of an synthetic alpha-synuclein induced cellular model reveals the expression profile of long non-coding RNA in Parkinson’s disease. Brain Res 1678:384–396
Mahfouz A, Huisman SMH, Lelieveldt BPF, Reinders MJT (2017) Brain transcriptome atlases: a computational perspective. Brain Struct Funct 222:1557–1580
Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R et al (2018) Developmental diversification of cortical inhibitory interneurons. Nature 555:457–462
Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105:716–721
Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L et al (2014) Transcriptional landscape of the prenatal human brain. Nature 508:199–206
Molnar Z, Kaas JH, de Carlos JA, Hevner RF, Lein E et al (2014) Evolution and development of the mammalian cerebral cortex. Brain Behav Evol 83:126–139
Mure LS, Le HD, Benegiamo G, Chang MW, Rios L et al (2018) Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359:eaao0318
Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I et al (2016) DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 44:D231-238
Paul Y, Thomas S, Patil V, Kumar N, Mondal B et al (2018) Genetic landscape of long noncoding RNA (lncRNAs) in glioblastoma: identification of complex lncRNA regulatory networks and clinically relevant lncRNAs in glioblastoma. Oncotarget 9:29548–29564
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369
Qin N, Tong GF, Sun LW, Xu XL (2017) Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol Res 25:1471–1478
Qu S, Liu Z, Yang X, Zhou J, Yu H et al (2018) The emerging functions and roles of circular RNAs in cancer. Cancer Lett 414:301–309
Rao J, Cheng X, Zhu H, Wang L, Liu L (2018) Circular RNA-0007874 (circMTO1) reverses chemoresistance to temozolomide by acting as a sponge of microRNA-630 in glioblastoma. Cell Biol Int. https://doi.org/10.1002/cbin.11080
Regev A, Teichmann SA, Lander ES, Amit I, Benoist C et al (2017) The human cell atlas. Elife 6:e27041
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885
Sarkar SN, Russell AE, Engler-Chiurazzi EB, Porter KN, Simpkins JW (2019) MicroRNAs and the genetic nexus of brain aging, neuroinflammation, neurodegeneration, and brain trauma. Aging Dis 10:329–352
Schaukowitch K, Kim TK (2014) Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience 264:25–38
Schwalb B, Michel M, Zacher B, Fruhauf K, Demel C et al (2016) TT-seq maps the human transient transcriptome. Science 352:1225–1228
Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF et al (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353:78–82
Straniero L, Rimoldi V, Samarani M, Goldwurm S, Di Fonzo A et al (2017) The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci Rep 7:12702
Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL et al (2013) Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41:D996-d1008
Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T et al (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19:335–346
Wang JB, Liu FH, Chen JH, Ge HT, Mu LY et al (2017) Identifying survival-associated modules from the dysregulated triplet network in glioblastoma multiforme. J Cancer Res Clin Oncol 143:661–671
Wang O, Huang Y, Wu H, Zheng B, Lin J et al (2018) LncRNA LOC728196/miR-513c axis facilitates glioma carcinogenesis by targeting TCF7. Gene 679:119–125
Wei R, Zhang L, Hu W, Wu J, Zhang W (2019) Long non-coding RNA AK038897 aggravates cerebral ischemia/reperfusion injury via acting as a ceRNA for miR-26a-5p to target DAPK1. Exp Neurol 314:100–110
Wilson JA, Prow NA, Schroder WA, Ellis JJ, Cumming HE et al (2017) RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog 13:e1006155
Wu W, Yu T, Wu Y, Tian W, Zhang J et al (2019) The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res 38:133
Xiao S, Wang R, Wu X, Liu W, Ma S (2018) The long noncoding RNA TP73-AS1 interacted with miR-124 to modulate glioma growth by targeting inhibitor of apoptosis-stimulating protein of p53. DNA Cell Biol 37:117–125
Xu C, He T, Li Z, Liu H, Ding B (2017) Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother 95:1504–1513
Xu N, Liu B, Lian C, Doycheva DM, Fu Z et al (2018) Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis 9:1139
Xue JY, Huang C, Wang W, Li HB, Sun M et al (2018) HOXA11-AS: a novel regulator in human cancer proliferation and metastasis. Onco Targets Ther 11:4387–4393
Yan H, Rao J, Yuan J, Gao L, Huang W et al (2017) Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis 8:3211
Yang BY, Meng Q, Sun Y, Gao L, Yang JX (2018) Long non-coding RNA SNHG16 contributes to glioma malignancy by competitively binding miR-20a-5p with E2F1. J Biol Regul Homeost Agents 32:251–261
Yao P, Lin P, Gokoolparsadh A, Assareh A, Thang MW et al (2015) Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat Neurosci 18:1168–1174
Zapata JC, Salvato MS (2015) Genomic profiling of host responses to Lassa virus: therapeutic potential from primate to man. Future Virol 10:233–256
Zhang S, Zhu D, Li H, Li H, Feng C et al (2017) Characterization of circRNA-associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol Ther 25:2053–2061
Zhang H, Li D, Zhang Y, Li J, Ma S et al (2018a) Knockdown of lncRNA BDNF-AS suppresses neuronal cell apoptosis via downregulating miR-130b-5p target gene PRDM5 in acute spinal cord injury. RNA Biol 15:1071–1080
Zhang L, Luo X, Chen F, Yuan W, Xiao X et al (2018b) LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem 119:5460–5472
Zhang R, Jin H, Lou F (2018c) The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J Cell Biochem 119:3007–3016
Zhang L, Xue Z, Yan J, Wang J, Liu Q et al (2019) LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif 52:e12573
Zhong S, Zhang S, Fan X, Wu Q, Yan L et al (2018) A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555:524–528
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81860644, 81560596, and 31560051), and Joint Special Project of Yunnan Science and Technology Department and Kunming Medical University (2019FE001-002)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ding, Z., Sun, L., Yang, C. et al. Exploring the secrets of brain transcriptional regulation: developing methodologies, recent significant findings, and perspectives. Brain Struct Funct 226, 313–322 (2021). https://doi.org/10.1007/s00429-021-02230-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-021-02230-x