Abstract
TFE3/TFEB-rearranged renal cell carcinomas are characterized by translocations involving TFE3 and TFEB genes. Despite the initial description of typical morphology, their histological spectrum is wide, mimicking common subtypes of renal cell tumors. Thus, the diagnosis is challenging requiring the demonstration of the gene rearrangement, usually by FISH. However, this technique is limited in most laboratories and immunohistochemical TFE3/TFEB analysis is inconsistent. We sought to identify a useful immunohistochemical panel using the most common available markers to recognize those tumors. We performed an immunohistochemical panel comparing 27 TFE3-rearranged and 10 TFEB-rearranged renal cell carcinomas to the most common renal cell tumors (150 clear cell, 100 papillary, 50 chromophobe renal cell carcinomas, 18 clear cell papillary renal cell tumors, and 50 oncocytomas). When dealing with neoplasms characterized by cells with clear cytoplasm, CA9 is a helpful marker to exclude clear cell renal cell carcinoma. GATA3, AMACR, and CK7 are useful to rule out clear cell papillary renal cell tumor. CK7 is negative in TFE3/TFEB-rearranged renal cell carcinoma and positive in papillary renal cell carcinoma, being therefore useful in this setting. Parvalbumin and CK7/S100A1 respectively are of paramount importance when TFE3/TFEB-rearranged renal cell carcinoma resembles oncocytoma and chromophobe renal cell carcinoma. Moreover, in TFEB-rearranged renal cell carcinoma, cathepsin K and melanogenesis markers are constantly positive, whereas TFE3-rearranged renal cell carcinoma stains for cathepsin K in roughly half of the cases, HMB45 in 8% and Melan-A in 22%. In conclusion, since TFE3/TFEB-rearranged renal cell carcinoma may mimic several histotypes, an immunohistochemical panel to differentiate them from common renal cell tumors should include cathepsin K, CA9, CK7, and parvalbumin.
Similar content being viewed by others
Introduction
Molecular classification of renal cell carcinoma has been evolving in the last decades, with emerging of new entities and new genetic characteristics. The forthcoming Word Health Organization (WHO) includes TFE3-rearranged renal cell carcinoma and TFEB-rearranged renal cell carcinoma as separate entities differentiating from the previous one in which the term “MiT family translocation renal cell carcinoma” encompassed both tumor types [1].
Both tumors are frequently discovered during childhood [2]; however, they can affect older patients as well and are both characterized by a translocation involving one of the MiT subfamily transcription factor genes. Despite these common aspects, the two entities present several differences. TFE3-rearranged renal cell carcinoma, as the name indicates, harbors TFE3 gene translocation which fuses with one of several other genes, such as ASPL (ASPSCR1), PRCC, SFPQ, CLTC, PARP14, RBM10, NONO, and MED15 [3,4,5,6,7,8,9,10,11]. There is a slight female predominance and half of those tumors presented at an advanced stage [10, 12] and behaved aggressively [13]. Histologically, the most typical cases show a mixed papillary and nested pattern with a mixture of cells with clear and granular/oncocytic cytoplasm. Psammoma bodies are often present within the tumor and might be a useful morphologic clue [14]. In TFEB-rearranged renal cell carcinoma TFEB gene, located on chromosome 6, most commonly translocated to chromosome 11 where fused with the Alpha (MALAT1) gene [15] and for this reason was previously designated as t(6;11) renal cell carcinoma. However, as TFE3-rearranged renal cell carcinoma, other gene fusion partners have been recently detected (COL21A1, CADM2, KHDRBS2, ACTB, EWSR1, CTLC, and NEAT1) [16,17,18,19,20], as well as tumors harboring TFEB gene amplification despite translocation [21,22,23,24,25,26,27,28]. There is no distinct gender predominance and most of them have an indolent clinical course [13]. Morphologically, the tumors are usually made up of a biphasic proliferation of large epithelioid clear and eosinophilic cells merged with aggregates of smaller cells gathered around spheres of basement membrane-derived material [29, 30].
Although these characteristic patterns can be striking in the classic cases, much more often, those tumors present heterogeneous architectural and cytological features, resembling the common subtypes of renal cell carcinoma. TFE3-rearranged renal cell carcinoma may show a solid, trabecular, or microcystic pattern, mimicking clear cell renal cell carcinoma, papillary renal cell carcinoma, and clear cell papillary renal cell tumor [2, 31,32,33]. On the other hand, in TFEB-rearranged renal cell carcinoma, a wide range of histological features has been reported, including extensive hyalinization, papillary architecture, clear cell morphology, or eosinophilic appearance mimicking papillary renal cell carcinoma, clear cell renal cell carcinoma, oncocytoma, chromophobe renal cell carcinoma, and epithelioid angiomyolipoma/pure epithelioid PEComa [29, 34, 35].
Due to the wide spectrum of morphology observed, the diagnosis is not straightforward on hematoxylin and eosin slides, and the identification of TFE3 or TFEB gene translocation is required to reach the proper diagnosis. Although it could be argued to use TFE3 and TFEB immunostaining to demonstrate the translocation, the results are inconsistent due to the not infrequent false-positive and false-negative results [36]. For this reason, fluorescent in situ hybridization (FISH) analysis is currently considered the gold standard [36,37,38,39]. Recently, the expression of TRIM63 by RNA in situ hybridization (RNA-ISH) assay has been proposed as another diagnostic marker for TFE3 and TFEB-rearranged renal cell carcinoma [40] even if an external validation has not been performed so far. Nevertheless, either FISH or RNA-ISH techniques are limited in most laboratories. Conversely, immunohistochemistry is more commonly available, and pathologists are more familiar to use this tool for diagnostic purposes, especially immunohistochemical markers performed in their practice. However, the range of positive expression of an immunohistochemical marker can be broad and several cutoffs have been reported in the literature and used in clinical practice. For these reasons, in this study, we have performed a detailed immunohistochemical analysis of TFE3 and TFEB-rearranged renal cell carcinoma compared with the most common histotypes of renal cell neoplasms evaluating its usefulness, with different cutoffs, in the differential diagnosis and looking for an immunohistochemical panel using most available markers to recognize those tumors.
Materials and methods
Patients and samples
Twenty-seven TFE3-rearranged renal cell carcinomas and ten TFEB-rearranged renal cell carcinomas were demonstrated by FISH analysis (Fig. 1). One hundred and fifty clear cell renal cell carcinomas, one hundred papillary renal cell carcinomas, fifty oncocytomas, fifty chromophobe renal cell carcinomas, and eighteen clear cell papillary renal cell tumors were also retrieved from the archives of the Pathology Department of Verona University and Pederzoli Hospital, Peschiera del Garda, Verona. All slides were reviewed by three experienced pathologists (AC, MB, and GM).
Immunohistochemistry
Sections from tissue blocks of all the included types of renal cell carcinoma were immunohistochemically stained with the following antibodies: PAX8 (clone BC12; DSB), CD10 (clone 56C6, dilution 1:50; Novocastra), carbonic anhydrase 9 CA9 (polyclonal rabbit, dilution 1:1000; Abcam), cytokeratin 7 (clone RN7, dilution 1:100; Novocastra), alpha-methylacyl-CoA racemase AMACR (clone 13H7, dilution 1:25; Dako), S100A1 (clone M01, dilution 1:600; Abnova), parvalbumin (clone P19, dilution 1:500; Sigma), CD13 (clone 38C12, dilution 1:100; Novocastra), GATA3 (clone L50-823, dilution 1:150; BD Pharmingen), and cathepsin K (clone 3F9, dilution 1:2000; Abcam). To further analyze the immunohistochemical profile of MiT family translocation renal cell carcinomas, TFE3-rearranged-renal cell carcinomas and TFEB-rearranged renal cell carcinomas were also stained with HMB45 (dilution 1:30; Dako, Denmark), Melan-A (clone A103, dilution 1:50; Novocastra, UK), CD68 (clone PG-M1, dilution 1:50; Dako), cytokeratin 8–18 (clone 5D3, dilution 1:100; Novocastra), cytokeratin 20 (clone PW31, dilution 1:100; Novocastra), fumarate hydratase FH (clone J-13, dilution 1:50; Santa Cruz), and succinate dehydrogenase B SDHB (clone 21A11AE7, dilution 1:800, Abcam).
All samples were processed using a sensitive Bond Polymer Refine detection system in an automated Bond immunohistochemistry instrument (Leica Biosystems, Germany). The immunohistochemical expression of each marker for every tumor subtype was recorded. Concerning TFE3 and TFEB-rearranged renal cell carcinomas, three different cutoffs of expression were evaluated for each marker, respectively of 5%, 10%, and 20%.
Statistical analysis
Fisher’s exact test was used to compare categorical data for immunohistochemical characteristics for each of above mentioned three different cutoff levels. All P-values were based on a two-tailed hypothesis. The results were considered statistically significant if the P-value was less than 0.05.
Results
MiT family translocation renal cell carcinomas (TFE3 and TFEB-rearranged renal cell carcinomas)
The pathological features of twenty-seven TFE3-rearranged renal cell carcinomas [41] and ten TFEB-rearranged renal cell carcinomas [19, 27, 35, 41, 42] have already been reported. The immunohistochemical results are here further expanded and detailed in Table 1 recording the percentage of expression.
Cathepsin K was observed in the most of MiT family translocation renal cell carcinomas (66% and 63% using the threshold of 5% and 10% or 20% positive cells respectively). About TFE3-rearranged renal cell carcinomas, immunolabeling was observed in roughly half of the cases (54% and 50% using the threshold of 5% and 10% or 20% positive cells respectively) whereas all the eleven TFEB-rearranged renal cell carcinomas evaluated stained positive for cathepsin K.
Melanocytic markers, HMB45 and Melan-A, were respectively positive in 15 of 36 (42%) and 14 of 34 (41%) of MiT family translocation renal cell carcinomas using the 5% cutoff. The percentage of cases considered positive for HMB45 drastically decreased when the higher cutoff was used (17% and 11% using the threshold of 10% and 20% respectively) while the percentage of positive cases was similar for Melan-A regardless of the cutoff (38% and 29% using the threshold of 10% and 20% respectively). TFEB-rearranged renal cell carcinomas were constantly immunolabeled for Melan-A (100%, 100%, and 90% of cases using the threshold of 5%, 10%, and 20% positive cells respectively), making such a reliable marker for the identification of these tumors, and frequently for HMB45 (80%, 20%, and 10% of the cases using the threshold of 5%, 10%, and 20% positive cells respectively). The expression of Melan-A and HMB45 was significantly lower in TFE3-rearranged renal cell carcinoma compared with TFEB-rearranged renal cell carcinoma, even when a 5% cutoff was used (17% and 27% respectively).
Regarding the proximal tubular markers such as CD10 and CD13, immunolabeling for the former was observed in 26 of 33 (79%), 25 of 33 (76%), and 22 of 33 (67%) of the MiT family translocation renal cell carcinomas, with the respective increasing cutoffs. As for TFE3-rearranged renal cell carcinomas, among 23 tumors, CD10 stained positive in 22 of the cases (96%), regardless of the cutoff considered, whereas it was positive just in 4 (40%), 3 (30%), and none (0%) of all the ten cases of TFEB-rearranged renal cell carcinomas, respectively, using a 5%, 10%, and 20% positivity threshold. CD13 expression, instead, was found in 13 of 32 (41%) and 10 of 32 (31%) MiT family translocation renal cell carcinomas tested, respectively, with a 5% and both a 10% and 20% cutoff. Among TFE3-rearranged renal cell carcinomas, 11 of 23 tumors (48%), considering a 5% cutoff, and 9 of 23 tumors (39%), both with a 10% and a 20% threshold, were positive for CD13; a weak expression of the same marker was instead noticed for TFEB-rearranged renal cell carcinomas, with only 2 (22%) and 1 (11%) of 9 tumors staining positive, employing a 5% and both a 10% and 20% respectively considered.
As for the distal tubular markers such as GATA3 and parvalbumin, none of the MiT family translocation renal cell carcinomas retrieved was positive for the former. About parvalbumin expression, among 33 MiT family translocation renal cell carcinomas 12 (36%), using a 5% cutoff, and 7 (21%), both considering a 10% and 20% cutoff, labeled positive for it. While 5 of 22 TFE3-rearranged renal cell carcinomas showed positivity for such marker in more than 20% of the cells (22%), the remaining 17 cases were completely negative or patchy positive for it. Moreover, 7 of 10 (70%) and 2 of 10 (20%) TFEB-rearranged renal cell carcinomas revealed positive parvalbumin expression respectively considering a 5% threshold and both a 10% and a 20% threshold.
Considering other markers commonly evaluated when dealing with renal cell carcinomas, CA9 was expressed by 6 of 35 (17%), 2 of 35 (6%), and none of 35 (0%) MiT family translocation renal cell carcinomas, respectively, using a 5%, 10%, and 20% cutoff. Namely, whereas CA9 immunolabeling was found in 6 of 25 (24%) TFE3-rearranged renal cell carcinomas with a 5% threshold, in 2 of them (8%) and none of them (0%), respectively, referring to a 10% and 20% cutoff, all the TFEB-rearranged renal cell carcinomas were negative for such marker. Among the 35 MiT family translocation renal cell carcinomas tested, AMACR, instead, stained positive in 26 (74%), 19 (54%), and 18 cases (51%), regarding a 5%, 10%, and 20% cutoff respectively. Positive expression was found in 23 of 25 (92%), 19 of 25 (76%), and 18 of 25 (72%) TFE3-rearranged renal cell carcinomas, using the same thresholds. Nevertheless, only 3 of 10 (30%) TFEB-rearranged renal cell carcinomas were considered positive for AMACR when a 5% cutoff was used, while none of them showed positive staining in more than 10% of the cells.
Furthermore, S100A1 was typically positive and CK7 was usually negative regardless of the cutoff used either considering the overall MiT family translocation renal cell carcinomas or TFE3 and TFEB-rearranged renal cell carcinoma separately. Regarding CK8-18 immunolabeling instead, it was noticed in 19 of 34 (56%), 15 of 34 (44%), and 13 of 34 (38%) MiT family translocation renal cell carcinomas, using a 5%, 10%, and 20% cutoff respectively. While the expression is lower for TFE3-rearranged renal cell carcinomas (41%, 33%, and 29% respectively with a 5%, a 10%, and 20% threshold), most of TFEB-rearranged renal cell carcinomas labeled positive for CK8-18 (100%, 80%, and 60% respectively with 5%, 10%, and 20% threshold).
None of the MiT family translocation renal cell carcinomas considered expressed CD68 (PG-M1) neither CK20. SDHB and FH were retained in all the cases tested.
The immunohistochemical results of clear cell renal cell carcinoma, papillary renal cell carcinoma, clear cell papillary renal cell tumor, chromophobe renal cell carcinoma, and oncocytoma are detailed in supplementary Table 1.
Comparison of immunohistochemical markers and statistical relevance
MiT family translocation renal cell carcinoma versus clear cell renal cell carcinoma
Regardless of the cutoff used, in the differential diagnosis with clear cell renal cell carcinoma, a statistically significant correlation was found with MiT family translocation renal cell carcinomas and negative expression of CA9 (p = 0.0001) and CD13 (p = 0.0001), with MiT family translocation renal cell carcinomas and positive expression of AMACR (p = 0.0001), cathepsin K (p = 0.0001), and parvalbumin (p = 0.0001 with a 5% cutoff and p = 0.0007 with both a 10% and a 20% cutoff).
As far as TFE3-rearranged renal cell carcinomas were concerned, despite the cutoff taken into account, a negative expression of CA9 (p = 0.0001) and CD13 (p = 0.0003 with a 5% cutoff and p = 0.0001 with both a 10% and a 20% cutoff) statistically correlated with the diagnosis of such tumor subtype, as well as cathepsin K (p = 0.0001), AMACR (p = 0.0001), and parvalbumin (p = 0.0021) positivity.
About TFEB-rearranged renal cell carcinomas, finally, a statistical correlation of relevant levels was reported with negative expression of CD10 (p = 0.0002 with a 5% cutoff and p = 0.0001 with both a 10% and a 20% cutoff), CA9 (p = 0.0002 with both a 5% and a 10% cutoff and p = 0.0001 with both a 20% cutoff), and CD13 (p = 0.0001) and with cathepsin K (p = 0.0001) and parvalbumin positivity (p = 0.0001 with a 5% threshold and p = 0.0048 with both a 10% and a 20% threshold), no matter what the cutoff used was.
Useful tools
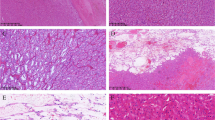
CD10 is not useful in the differential diagnosis between clear cell renal cell carcinoma and TFE3-rearranged renal cell carcinoma since both tumors are usually labeled for this marker, but it helps distinguish from TFEB-rearranged renal cell carcinoma. CA9 is usually negative either in TFE3 or TFEB-rearranged renal cell carcinoma whereas it is an important positive reliable marker in clear cell renal cell carcinoma. On the other hand, cathepsin K is positive in TFEB-rearranged renal cell carcinoma and half of TFE3-rearranged renal cell carcinoma while it is negative in clear cell renal cell carcinoma (Fig. 2).
A TFE3-rearranged renal cell carcinoma composed of cells with clear cytoplasm (A) and an example of clear cell renal cell carcinoma arranged in nests (B). Both tumors are positive for CD10 (C, D), whereas CA9 is negative in TFE3-rearranged renal cell carcinoma (E) typically expressed in clear cell carcinoma (F)
Suggested panel
Suggested panel is as follows: CA9 and cathepsin K.
MiT family translocation renal cell carcinoma versus clear cell papillary renal cell tumor
When referring to clear cell papillary renal cell tumor, there was a strong statistically significant correlation, despite the threshold of positivity considered, with MiT family translocation renal cell carcinomas and negative expression of CA9 (p = 0.0001), CK7 (p = 0.0001), and GATA3 (p = 0.0001), along with positive expression of CD10 (p = 0.0001 with both a 5% and 10% cutoff and p = 0.0011 with a 20% cutoff), AMACR (p = 0.0001 with both a 5% and 10% cutoff and p = 0.0002 with a 20% cutoff), and cathepsin K (p = 0.0001). Similar results were observed when TFE3-rearranged renal cell carcinoma was considered. However, in TFEB-rearranged renal cell carcinoma, CD10 and AMACR staining are not statistically correlated (p = 0.5 and p = 1 respectively).
Useful tools
GATA3, CK7, and CA9 are the most reliable markers in this differential diagnosis since are positive in clear cell papillary renal cell tumor and negative in TFE3 and TFEB-rearranged. CD10 is not useful in the differential diagnosis between clear cell papillary renal cell tumor and TFEB-rearranged renal cell carcinoma since both tumors are typically negative for this marker, but it helps distinguish from TFE3-rearranged renal cell carcinoma, usually positive. Cathepsin K is positive in TFEB-rearranged renal cell carcinoma and half of TFE3-rearranged renal cell carcinoma while it is negative in clear cell papillary renal cell tumor (Fig. 3).
A cystic TFE3-rearranged renal cell carcinoma lined by cells with clear cytoplasm (the inset underlining the nuclei oriented toward the lumen) (A), is negative for CK7 (C) and GATA3 (E). Conversely, a clear cell papillary renal cell tumor nearly entirely cystic (the inset highlighting a partially papillary architecture) (B), showing CK7 (D) and GATA3 (F) positivity
Suggested panel
Suggested panel is as follows: CA9, CK7, GATA3, and cathepsin K.
MiT family translocation renal cell carcinoma versus papillary renal cell carcinoma
As for the differential diagnosis with papillary renal cell carcinoma, despite what cutoff was used, a significant statistical correlation was noticed with MiT family translocation renal cell carcinomas and negative expression of CK7 (p = 0.0001), AMACR (p = 0.066 with a 5% cutoff and p = 0.0001 with both a 10% and 20% cutoff), and CD13 (p = 0.0001 with both a 5% and 10% cutoff and p = 0.0002 with a 20% cutoff) and with MiT family translocation renal cell carcinomas and positive expression of parvalbumin (p = 0.0001 with a 5% threshold and p = 0.0095 with a 10% and 20% threshold) and cathepsin K (p = 0.0001).
Speaking of TFE3-rearranged renal cell carcinomas, regardless the cutoff, a negative expression of only CK7 (p = 0.0001) and CD13 (p = 0.0018 with a 5% cutoff, p = 0.0002 with a 10% cutoff and p = 0.0001 with a 20% one) was of statistically significant value, as well as positive immunolabeling for CD10 (p = 0.0002), parvalbumin (p = 0.0016), and cathepsin K (p = 0.0001). Furthermore, negative expression of CA9 (p = 0.017) and AMACR (p = 0.0045), both considering a 20% positivity cutoff, and positive staining for S100A1 (p = 0.0282), with the lowest cutoff of 5%, was significantly strong alike.
A statistically relevant correlation was also noted between TFEB-rearranged renal cell carcinomas and negativity for CK7 (p = 0.0001), AMACR (p = 0.0001), and CD13 (p = 0.0003 with a 5% threshold and p = 0.0001 with both a 10% and a 20% threshold) along with their positive expression of cathepsin K (p = 0.0001), no matter what cutoff value was considered.
Useful tools
AMACR is not useful in the differential diagnosis between papillary renal cell carcinoma and TFE3-rearranged renal cell carcinoma since both tumors are usually labeled for this marker, but it helps distinguish from TFEB-rearranged renal cell carcinoma. CK7 is usually negative either in TFE3 or TFEB-rearranged renal cell carcinoma whereas it is an important positive reliable marker in papillary renal cell carcinoma. On the other hand, cathepsin K is positive in TFEB-rearranged renal cell carcinoma and half of TFE3-rearranged renal cell carcinoma while it is negative in papillary renal cell carcinoma (Fig. 4).
TFE3-rearranged renal cell carcinoma with papillary architecture (A) and papillary renal cell carcinoma (B). Both tumors expressed AMACR (C, D) which is not useful in this differential diagnosis. Staining for CK7 is negative in TFE3-rearranged renal cell carcinoma (E) but positive in papillary renal cell carcinoma (F)
Suggested panel
Suggested panel is as follows CK7 and cathepsin K.
MiT family translocation renal cell carcinoma versus chromophobe renal cell carcinoma
In cases with histological features recalling that of chromophobe carcinomas, negative expression of CK7 (p = 0.0001) and parvalbumin (p = 0.0001), along with immunolabeling for CD10 (p = 0.0001), AMACR (p = 0.0001), S100A1 (p = 0.0001), CD13 (p = 0.0001 with a 5% threshold and p = 0.0008 with both a 10% and 20% threshold), and cathepsin K (p = 0.0001) were statistically consistent with the diagnosis of MiT family translocation renal cell carcinomas, all these results observed concerning each of the above-mentioned positivity cutoffs.
TFE3-rearranged renal cell carcinomas revealed the same results. In addition, interestingly, positive staining for CA9 was also found to be statistically relevant, but only when a 5% positivity cutoff was considered (p = 0.0239).
Finally, as far as TFEB-rearranged renal cell carcinomas were concerned, negativity for CK7 (p = 0.0159 with both a 5% and 10% cutoff and p = 0.0014 with a 20% one) and parvalbumin (p = 0.0183 with a 5% threshold and p = 0.0001 with both a 10% and 20% one) as well as positive expression of S100A1 (p = 0.0001) and cathepsin K (p = 0.0001) statistically correlated with the diagnosis of such tumor subtype, despite the cutoff chosen. On the other hand, no relevant statistical correlation was found with immunohistochemical expression of either CD10, AMACR, or CD13, in contrast with the data observed with TFE3-rearranged renal cell carcinomas.
Useful tools
CK7, S100A1, and parvalbumin are helpful immunohistochemical markers in the differential diagnosis between chromophobe renal cell carcinoma and TFE3/TFEB-rearranged renal cell carcinoma. Expression of CK7 and parvalbumin along with the absence of S100A1 is characteristic of chromophobe renal cell carcinoma, whereas strong and diffuse labeling of S100A1 along with the under-expression of CK7 and parvalbumin are typical of TFE3/TFEB-rearranged renal cell carcinoma. Cathepsin K is positive in TFEB-rearranged renal cell carcinoma and half of TFE3-rearranged renal cell carcinoma while it is negative in chromophobe renal cell carcinoma.
Suggested panel
Suggested panel is as follows: CK7, S100A1, parvalbumin, and cathepsin K.
MiT family translocation renal cell carcinoma versus oncocytoma
Negative expression of parvalbumin (p = 0.0001) and positive staining for CD10 (p = 0.0001 with a 5% cutoff, p = 0.0004 with a 10% cutoff, and p = 0.0070 with a 20% cutoff), AMACR (p = 0.0001), CD13 (p = 0.0001), and cathepsin K (p = 0.0001) were strongly statistically consistent with the diagnosis of MiT family translocation renal cell carcinoma despite what positivity threshold was used. Similar results were also noted for negative expression of S100A1 (p = 0.0305) and CK7 (p = 0.0364), respectively referring to a 10% and 20% cutoff.
The analysis from TFE3-rearranged renal cell carcinomas data revealed similar results, whereas, regarding TFEB-rearranged renal cell carcinomas, cathepsin K positive staining was the only immunohistochemical result of statistical significance for all the positivity thresholds considered (p = 0.0001) and a statistical correlation was also collected with parvalbumin negativity (p = 0.0001), even though only with a 5% and 10% positivity threshold.
Useful tools
Cathepsin K and parvalbumin are the two useful markers in the differential diagnosis between oncocytoma and TFE3/TFEB-rearranged renal cell carcinoma (Fig. 5).
Suggested panel
Suggested panel is as follows: parvalbumin and cathepsin K.
In Tables 2, 3, and 4, the comparison for the immunohistochemical markers with the p-value among MiT family translocation renal cell carcinomas and renal cell tumors is shown, respectively. using a 5%, 10%, and 20% positivity threshold.
Discussion
Despite initially considered rare tumors, TFE3 and TFEB-rearranged renal cell carcinoma represent 1–4% of renal cell carcinomas diagnosed among adults [13]. Proper identification of these tumor types is challenging since histological features may be ambiguous and often overlap with other more common types of renal cell neoplasm. In general, TFE3 and TFEB-rearranged renal cell carcinomas ought to be considered in the differential diagnosis, especially in young patients, every time pathologists have to deal with a renal tumor showing unusual microscopic findings [13, 29]. To support the diagnosis, TFE3/TFEB gene translocation should be demonstrated by FISH break-apart assay or the gene fusion identified by RNA sequencing [43]. Nevertheless, these techniques are not available in all laboratories and a thorough evaluation of their immunophenotype might be worth it for correctly identifying them. Moreover, in the last years, increasing use of immunohistochemistry has been observed due to the new clinic-pathologic entities identified.
Several studies addressed the immunohistochemical profile to aid in the classcarcinoma [44,45,46]. Staining for cathepsin K, CA9, CK7, and HMB45 has been claimed as helpful for this purpose. Basically, translocation renal cell carcinoma is labeled for cathepsin K and HMB45 while it is negative for CA9 and CK7. Moreover, previous manuscripts mainly focused on the differential diagnosis between TFE3-rearranged renal cell carcinoma and clear cell renal cell carcinoma [44, 46]. As previously pointed out, TFE3-rearranged renal cell carcinoma is likely to be misdiagnosed as clear cell renal cell carcinoma if a restricted immunohistochemical panel is applied. In this scenario, CD10 is not helpful regardless of the threshold of positivity considered, whereas performing cathepsin K and CA9 is useful in sorting the diagnostic quandary out. However, staining for CD10 has an important value in the differential diagnosis with TFEB-rearranged renal cell carcinoma, along with cathepsin K and CA9.
Besides clear cell renal cell carcinoma, other tumors can be misclassified. TFE3-rearranged renal cell carcinoma may also resemble clear cell papillary renal cell tumor. Those neoplasms usually labeled for CK7, CA9, and GATA3, not expressed in TFE3-rearranged renal cell carcinomas, which are instead commonly positive for cathepsin K. Either TFE3-rearranged renal cell carcinomas or TFEB-rearranged renal cell carcinomas demonstrating papillary architecture can be confused with papillary renal cell carcinoma. While AMACR is not useful in the differential diagnosis with TFE3-rearranged renal cell carcinomas, being positive in both tumors, such marker is usually under-expressed in TFEB-rearranged renal cell carcinomas. Of course, staining for CK7 favors papillary renal cell carcinoma, confirming its usefulness in distinguishing papillary renal cell carcinoma from TFE3 and TFEB-rearranged renal cell carcinomas. When TFE3 and TFEB-rearranged renal cell carcinomas exhibit extensive eosinophilic features, they can mimic oncocytoma and the eosinophilic variant of chromophobe renal cell carcinoma. To distinguish them, cathepsin K and parvalbumin are very helpful, in addition to CK7 and S100A1 for chromophobe renal cell carcinoma [47, 48]. Nevertheless, cathepsin K is expressed in other oncocytic neoplasms, such as the recently described eosinophilic solid and cystic renal cell carcinoma [49, 50]. These tumors may show overlapping morphological and immunohistochemical features with MiT family translocation renal cell carcinoma, especially with TFEB-rearranged renal cell carcinoma, both positive for cathepsin K and Melan-A. Although not addressed in the present manuscript, eosinophilic solid and cystic renal cell carcinomas immunolabeled for CK20 [51, 52], unlike TFEB-rearranged renal cell carcinoma, which has been claimed as a reliable marker in the differential diagnosis. However, conflicting data have been reported about the role of CK20 as a key diagnostic marker [53], so molecular analysis by FISH assay looking for TFEB gene alterations is still warranted in the most controversial cases.
Another important aspect of this study is the proper cutoff employed to define an immunohistochemical marker as positive since positive/negative results can be considered differently among pathologists. As different thresholds of positivity have been reported, it is important to establish if there are variances in results using several cutoffs of expression to consider an immunohistochemical marker as positive or negative. As expected, the threshold for positivity established is important for HMB45 and Melan-A. Even a patchy expression should be considered a positive result, especially for TFE3-rearranged renal cell carcinoma in which melanogenesis markers are less consistent. On the other hand, the different percentages (> 5%, > 10%, > 20%) of neoplastic cells labeling for cathepsin K do not change the result since the expression is usually strong and diffuse [54, 55]. Interestingly, some p-values of the different immunohistochemical markers vary based on the different percentages (> 5%, > 10%, > 20%). For instance, in the differential diagnosis between papillary renal cell carcinoma and TFE3-rearranged renal cell carcinoma, AMACR results are statistically significant when used 20% of positivity as a threshold whereas its expression is not significant with lower cutoffs, supporting the hypothesis that it is better to consider a lower percentage of positivity. On the other side, S100A1 seems an important marker in the differential diagnosis between TFEB-rearranged renal cell carcinoma and papillary renal cell carcinoma (p = 0.0152) using 5% and 10% of positivity whereas it is not significant with higher cutoffs, suggesting to use the high threshold of positivity. Overall, aside from melanogenesis markers and AMACR, in clinical practice, it is better to use a higher threshold to consider a positive immunohistochemical marker.
Finally, the study highlighted the importance, also from an immunohistochemical point of view, to maintain TFE3 and TFEB-rearranged renal cell carcinoma as distinct entities, as recommended by the forthcoming WHO classification of renal tumors. When grouped together under the heading MiT family translocation renal cell carcinoma, the value of some immunohistochemical markers changed dramatically. For instance, staining for CD10 which is usually observed in TFE3-rearranged renal cell carcinoma and under-expressed in TFEB-rearranged renal cell carcinoma seems significant in the differential diagnosis of MiT family translocation renal cell carcinoma and clear cell renal cell carcinoma. However, as abovementioned, CD10 is not useful in this differential diagnosis when we are dealing with TFE3-rearranged renal cell carcinoma.
In conclusion, the threshold to define a positive immunohistochemical result is not univocal but depends on the immunohistochemical marker considered. For instance, any expression of melanogenesis markers should be considered positive. A panel of immunohistochemistry markers useful to distinguish TFE3/TFEB-rearranged renal cell carcinoma from other common renal cell neoplasms should include cathepsin K, CA9, CK7, and parvalbumin (Table 5). In this scenario, tumors with equivocal morphological features and doubtful immunoprofile should be analyzed by FISH, which reliably detect the most common rearrangements. However, subtle TFE3 gene inversions, such as RBM10 and NONO gene fusions, may lead to false negative results by FISH and require further molecular analysis.
References
Moch H, Humphrey PA, Ulbright TM, Reuter VE (2016) WHO classification of tumours of the urinary system and male genital organs, 4th edn. IARC press, Lyon, France
Sukov WR, Hodge JC, Lohse CM et al (2012) TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am J Surg Pathol 36:663–670. https://doi.org/10.1097/PAS.0b013e31824dd972
Argani P, Antonescu CR, Illei PB et al (2001) Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol 159:179–192. https://doi.org/10.1016/S0002-9440(10)61684-7
Argani P, Antonescu CR, Couturier J et al (2002) PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am J Surg Pathol 26:1553–1566. https://doi.org/10.1097/00000478-200212000-00003
Xia Q-Y, Wang X-T, Zhan X-M et al (2017) Xp11 translocation renal cell carcinomas (RCCs) with RBM10-TFE3 gene fusion demonstrating melanotic features and overlapping morphology with t(6;11) RCC: interest and diagnostic pitfall in detecting a paracentric inversion of TFE3. Am J Surg Pathol 41:663–676. https://doi.org/10.1097/PAS.0000000000000837
Xia Q-Y, Wang Z, Chen N et al (2017) Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod Pathol 30:416–426. https://doi.org/10.1038/modpathol.2016.204
Argani P, Lui MY, Couturier J et al (2003) A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23). Oncogene 22:5374–5378. https://doi.org/10.1038/sj.onc.1206686
Clark J, Lu YJ, Sidhar SK et al (1997) Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 15:2233–2239. https://doi.org/10.1038/sj.onc.1201394
Pivovarcikova K, Grossmann P, Alaghehbandan R et al (2017) TFE3-fusion variant analysis defines specific clinicopathologic associations amog Xp11 translocation cancers. Am J Surg Pathol 41:138–140
Camparo P, Vasiliu V, Molinie V et al (2008) Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol 32:656–670. https://doi.org/10.1097/PAS.0b013e3181609914
Pei J, Cooper H, Flieder DB et al (2019) NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma. Mod Pathol 32:710–716. https://doi.org/10.1038/s41379-018-0191-7
Wu A, Kunju LP, Cheng L, Shah RB (2008) Renal cell carcinoma in children and young adults: analysis of clinicopathological, immunohistochemical and molecular characteristics with an emphasis on the spectrum of Xp11.2 translocation-associated and unusual clear cell subtypes. Histopathology 53:533–544. https://doi.org/10.1111/j.1365-2559.2008.03151.x
Caliò A, Segala D, Munari E et al (2019) MiT family translocation renal cell carcinoma: from the early descriptions to the current knowledge. Cancers (Basel) 11:1110. https://doi.org/10.3390/cancers11081110
Argani P (2021) Translocation carcinomas of the kidney. Genes Chromosomes Cancer. https://doi.org/10.1002/gcc.23007
Argani P, Hawkins A, Griffin CA et al (2001) A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol 158:2089–2096. https://doi.org/10.1016/S0002-9440(10)64680-9
Durinck S, Stawiski EW, Pavía-Jiménez A et al (2015) Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet 47:13–21. https://doi.org/10.1038/ng.3146
Xia Q-Y, Wang X-T, Fang R et al (2020) Clinicopathologic and molecular analysis of the TFEB fusion variant reveals new members of TFEB translocation renal cell carcinomas (RCCs): expanding the genomic spectrum. Am J Surg Pathol 44:477–489. https://doi.org/10.1097/PAS.0000000000001408
Malouf GG, Su X, Yao H et al (2014) Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin cancer Res an Off J Am Assoc Cancer Res 20:4129–4140. https://doi.org/10.1158/1078-0432.CCR-13-3036
Caliò A, Harada S, Brunelli M et al (2021) TFEB rearranged renal cell carcinoma. A clinicopathologic and molecular study of 13 cases. Tumors harboring MALAT1-TFEB, ACTB-TFEB, and the novel NEAT1-TFEB translocations constantly express PDL1. Mod Pathol 34:842–850. https://doi.org/10.1038/s41379-020-00713-6
Linehan WM, Spellman PT, Ricketts CJ et al (2016) Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 374:135–145. https://doi.org/10.1056/NEJMoa1505917
Skala SL, Xiao H, Udager AM et al (2018) Detection of 6 TFEB-amplified renal cell carcinomas and 25 renal cell carcinomas with MITF translocations: systematic morphologic analysis of 85 cases evaluated by clinical TFE3 and TFEB FISH assays. Mod Pathol 31:179–197. https://doi.org/10.1038/modpathol.2017.99
Lilleby W, Vlatkovic L, Meza-Zepeda LA et al (2015) Translocational renal cell carcinoma (t(6;11)(p21;q12) with transcription factor EB (TFEB) amplification and an integrated precision approach: a case report. J Med Case Rep 9:281. https://doi.org/10.1186/s13256-015-0749-7
Argani P, Reuter VE, Zhang L et al (2016) TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am J Surg Pathol 40:1484–1495. https://doi.org/10.1097/PAS.0000000000000720
Williamson SR, Grignon DJ, Cheng L et al (2017) Renal cell carcinoma with chromosome 6p amplification including the TFEB gene: a novel mechanism of tumor pathogenesis? Am J Surg Pathol 41:287–298. https://doi.org/10.1097/PAS.0000000000000776
Gupta S, Johnson SH, Vasmatzis G et al (2017) TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod Pathol 30:998–1012. https://doi.org/10.1038/modpathol.2017.24
Kammerer-Jacquet S-F, Gandon C, Dugay F et al (2022) Comprehensive study of nine novel cases of TFEB-amplified renal cell carcinoma: an aggressive tumour with frequent PDL1 expression. Histopathology. https://doi.org/10.1111/his.14683
Caliò A, Brunelli M, Segala D et al (2019) VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod Pathol 32:258–268. https://doi.org/10.1038/s41379-018-0128-1
Mendel L, Ambrosetti D, Bodokh Y et al (2018) Comprehensive study of three novel cases of TFEB-amplified renal cell carcinoma and review of the literature: evidence for a specific entity with poor outcome. Genes Chromosomes Cancer 57:99–113. https://doi.org/10.1002/gcc.22513
Argani P (2015) MiT family translocation renal cell carcinoma. Semin Diagn Pathol 32:103–113. https://doi.org/10.1053/j.semdp.2015.02.003
Petersson F, Vaněček T, Michal M et al (2012) A distinctive translocation carcinoma of the kidney; “rosette forming”, t(6;11), HMB45-positive renal tumor: a histomorphologic, immunohistochemical, ultrastructural, and molecular genetic study of 4 cases. Hum Pathol 43:726–736. https://doi.org/10.1016/j.humpath.2011.07.001
Ross H, Martignoni G, Argani P (2012) Renal cell carcinoma with clear cell and papillary features. Arch Pathol Lab Med 136:391–399. https://doi.org/10.5858/arpa.2011-0479-RA
Wang X-T, Xia Q-Y, Ye S-B et al (2018) RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations. Mod Pathol 31:1346–1360. https://doi.org/10.1038/s41379-018-0051-5
Hayes M, Peckova K, Martinek P et al (2015) Molecular-genetic analysis is essential for accurate classification of renal carcinoma resembling Xp11.2 translocation carcinoma. Virchows Arch 466:313–322. https://doi.org/10.1007/s00428-014-1702-7
Williamson SR, Eble JN, Palanisamy N (2017) Sclerosing TFEB-rearrangement renal cell carcinoma: a recurring histologic pattern. Hum Pathol 62:175–179. https://doi.org/10.1016/j.humpath.2016.10.024
Caliò A, Brunelli M, Segala D, Pedron S, Tardanico R, Remo A, Gobbo S, Meneghelli E, Doglioni C, Hes O, Zampini C, Argani P, Martignoni G (2018) t(6;11) renal cell carcinoma: a study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod Pathol 31(3):474–487. https://doi.org/10.1038/modpathol.2017
Green WM, Yonescu R, Morsberger L et al (2013) Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol 37:1150–1163. https://doi.org/10.1097/PAS.0b013e31828a69ae
Kim SH, Choi Y, Jeong HY et al (2011) Usefulness of a break-apart FISH assay in the diagnosis of Xp11.2 translocation renal cell carcinoma. Virchows Arch 459:299–306. https://doi.org/10.1007/s00428-011-1127-5
Rao Q, Williamson SR, Zhang S et al (2013) TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol 37:804–815. https://doi.org/10.1097/PAS.0b013e31827e17cb
Argani P, Laé M, Hutchinson B et al (2005) Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol 29:230–240. https://doi.org/10.1097/01.pas.0000146007.54092.37
Wang X-M, Zhang Y, Mannan R et al (2021) TRIM63 is a sensitive and specific biomarker for MiT family aberration-associated renal cell carcinoma. Mod Pathol 34:1596–1607. https://doi.org/10.1038/s41379-021-00803-z
Caliò A, Brunelli M, Segala D et al (2020) Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: investigating prognostic markers and therapy targets. Pathology 52:297–309. https://doi.org/10.1016/j.pathol.2019.11.006
Pecciarini L, Cangi MG, Lo Cunsolo C et al (2007) Characterization of t(6;11)(p21;q12) in a renal-cell carcinoma of an adult patient. Genes Chromosomes Cancer 46:419–426. https://doi.org/10.1002/gcc.20422
Harada S, Caliò A, Janowski KM et al (2021) Diagnostic utility of one-stop fusion gene panel to detect TFE3/TFEB gene rearrangement and amplification in renal cell carcinomas. Mod Pathol 34:2055–2063. https://doi.org/10.1038/s41379-021-00858-y
Argani P, Hicks J, De Marzo AM et al (2010) Xp11 translocation renal cell carcinoma (RCC): extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol 34:1295–1303. https://doi.org/10.1097/PAS.0b013e3181e8ce5b
Smith NE, Illei PB, Allaf M et al (2014) t(6;11) renal cell carcinoma (RCC): expanded immunohistochemical profile emphasizing novel RCC markers and report of 10 new genetically confirmed cases. Am J Surg Pathol 38:604–614. https://doi.org/10.1097/PAS.0000000000000203
Yang B, Duan H, Cao W et al (2019) Xp11 translocation renal cell carcinoma and clear cell renal cell carcinoma with TFE3 strong positive immunostaining: morphology, immunohistochemistry, and FISH analysis. Mod Pathol 32:1521–1535. https://doi.org/10.1038/s41379-019-0283-z
Caliò A, Ammendola S, Brunelli M et al (2021) Parvalbumin immunohistochemical expression in the spectrum of perivascular epithelioid cell (PEC) lesions of the kidney. Virchows Arch 478:785–791. https://doi.org/10.1007/s00428-020-02856-y
Martignoni G, Brunelli M, Gobbo S et al (2007) Role of molecular markers in diagnosis and prognosis of renal cell carcinoma. Anal Quant Cytol Histol 29:41–49
Trpkov K, Abou-Ouf H, Hes O et al (2017) Eosinophilic solid and cystic renal cell carcinoma (ESC RCC): further morphologic and molecular characterization of ESC RCC as a Distinct entity. Am J Surg Pathol 41:1299–1308. https://doi.org/10.1097/PAS.0000000000000838
Caliò A, Brunelli M, Gobbo S et al (2021) Cathepsin K: a novel diagnostic and predictive biomarker for renal tumors. Cancers (Basel) 13:2441. https://doi.org/10.3390/cancers13102441
Trpkov K, Hes O, Bonert M et al (2016) Eosinophilic, solid, and cystic renal cell carcinoma: clinicopathologic study of 16 unique, sporadic neoplasms occurring in women. Am J Surg Pathol 40:60–71. https://doi.org/10.1097/PAS.0000000000000508
Munari E, Settanni G, Caliò A et al (2022) TSC loss is a clonal event in eosinophilic solid and cystic renal cell carcinoma: a multiregional tumor sampling study. Mod Pathol 35:376–385. https://doi.org/10.1038/s41379-021-00816-8
Lobo J, Rechsteiner M, Helmchen BM et al (2022) Eosinophilic solid and cystic renal cell carcinoma and renal cell carcinomas with TFEB alterations: a comparative study. Histopathology 81:32–43. https://doi.org/10.1111/his.14663
Martignoni G, Gobbo S, Camparo P et al (2011) Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod Pathol 24:1313–1319. https://doi.org/10.1038/modpathol.2011.93
Martignoni G, Pea M, Gobbo S et al (2009) Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol 22:1016–1022. https://doi.org/10.1038/modpathol.2009.58
Funding
Open access funding provided by Università degli Studi di Verona within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
Conceptualization: AC and GM. Methodology: AC and GM. Formal analysis and investigation: AC and SM. Writing — original draft preparation: AC and GM. Writing — review and editing: MB, SP, SCP, DS, EB, SG, GN, and GM. Supervision: AC and GM.
Corresponding author
Ethics declarations
Ethics approval
All procedures performed in our study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the declaration of Helsinki.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caliò, A., Marletta, S., Brunelli, M. et al. TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms. Virchows Arch 481, 877–891 (2022). https://doi.org/10.1007/s00428-022-03380-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-022-03380-x