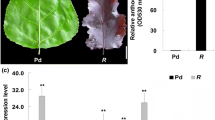
Abstract
Main conclusion
Two TT2-type MYB transcription factors identified from tetraploid cotton are involved in regulating proanthocyanidin biosynthesis, providing new strategies for engineering condensed tannins in crops.
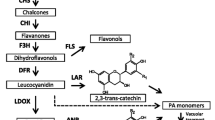
Proanthocyanidins (PAs), also known as condensed tannins, are important secondary metabolites involved in stress resistance in plants, and are health supplements that help to reduce cholesterol levels. As one of the most widely grown crops in the world, cotton provides the majority of natural fabrics and is a supplemental food for ruminant animals. The previous studies have suggested that PAs present in cotton are a major contributor to fiber color. However, the biosynthesis of PAs in cotton still remains to be elucidated. AtTT2 (transparent testa 2) is a MYB family transcription factor from Arabidopsis that initiates the biosynthesis of PAs by inducing the expression of multiple genes in the pathway. In this study, we isolated two R2R3-type MYB transcription factors from Gossypium hirsutum that are homologous to AtTT2. Expression analysis showed that both genes were expressed at different levels in various cotton tissues, including leaf, seed coat, and fiber. Protoplast transactivation assays revealed that these two GhMYBs were able to activate promoters of genes encoding enzymes in the PA biosynthesis pathway, namely anthocyanidin reductase and leucoanthocyanidin reductase. Complementation experiments showed that both of the GhMYBs were able to recover the transparent testa seed coat phenotype of the Arabidopsis tt2 mutant by restoring PA biosynthesis. Ectopic expression of either of the two GhMYBs in Medicago truncatula hairy roots increased the contents of anthocyanins and PAs compared to control lines expressing the GUS gene, and expression levels of MtDFR, MtLAR, and MtANR were also elevated in lines expressing GhMYBs. Together, these data provide new insights into engineering condensed tannins in cotton.








Similar content being viewed by others
Abbreviations
- PA:
-
Proanthocyanidin
- ANR:
-
Anthocyanidin reductase
- LAR:
-
Leucoanthocyanidin reductase
- DMACA:
-
Dimethylaminocinnamaldehyde
References
Albert S, Delseny M, Devic M (1997) BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J 11:289–299
Aron PM, Kennedy JA (2008) Flavan-3-ols: nature, occurrence and biological activity. Mol Nutr Food Res 52:79–104
Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102:2649–2654
Bell AA, El-Zik KM, Thaxton PM (1992) Chemistry, biological significance, and genetic control of proanthocyanidins in cotton (Gossypium spp.). In: Hemingway RW, Laks PE (eds) Plant polyphenols. Synthesis, properties, significance. Springer, USA, pp 571–595
Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167:1448–1470
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15:2514–2531
Dixon RA, Xie D-Y, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165:9–28
Dixon RA, Liu C, Jun JH (2013) Metabolic engineering of anthocyanins and condensed tannins in plants. Curr Opin Biotechnol 24:329–335
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Feng H, Tian X, Liu Y, Li Y, Zhang X, Jones BJ, Sun Y, Sun J (2013) Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton. PLoS One 8:e58820
Feng H, Li Y, Wang S, Zhang L, Liu Y, Xue F, Sun Y, Wang Y, Sun J (2014) Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). J Exp Bot 65:5759–5769
Gonzalez de Colmenares N, Ramírez-Martínez JR, Aldana JO, Ramos-Niño ME, Clifford MN, Pékerar S, Méndez B (1998) Isolation, characterisation and determination of biological activity of coffee proanthocyanidins. J Sci Food Agric 77:368–372
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325:412–421
Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S (2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol 159:1204–1220
Hinchliffe DJ, Condon BD, Thyssen G, Naoumkina M, Madison CA, Reynolds M, Delhom CD, Fang DD, Li P, McCarty J (2016) The GhTT2_A07 gene is linked to the brown colour and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibres. J Exp Bot 67:5461–5471. doi:10.1093/jxb/erw312
Hovav R, Faigenboim-Doron A, Kadmon N, Hu G, Zhang X, Gallagher JP, Wendel JF (2015) A transcriptome profile for developing seed of polyploid cotton. Plant Genome. doi:10.3835/plantgenome2014.08.0041
Ito C, Oki T, Yoshida T, Nanba F, Yamada K, Toda T (2013) Characterisation of proanthocyanidins from black soybeans: isolation and characterisation of proanthocyanidin oligomers from black soybean seed coats. Food Chem 141:2507–2512
Jun JH, Liu C, Xiao X, Dixon RA (2015) The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27:2860–2879
Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37:104–114
Kovinich N, Saleem A, Arnason JT, Miki B (2012) Identification of two anthocyanidin reductase genes and three red-brown soybean accessions with reduced anthocyanidin reductase 1 mRNA, activity, and seed coat proanthocyanidin amounts. J Agric Food Chem 60:574–584
Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann G, Treutter D, Schwab W, Hoffmann T, Szankowski I (2007) Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226:1243–1254
Li TC, Fan HH, Li ZP, Wei J, Lin Y, Cai YP (2012) The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiol Plant 34:813–818
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530
Liu Y, Shi Z, Maximova S, Payne MJ, Guiltinan MJ (2013) Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol 13:1–19
Liu C, Jun JH, Dixon RA (2014) MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol 165:1424–1439
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Mellway RD, Constabel CP (2009) Metabolic engineering and potential functions of proanthocyanidins in poplar. Plant Signal Behav 4:790–792
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145:601–615
Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105:14210–14215
Pang Y, Abeysinghe ISB, He J, He X, Huhman D, Mewan KM, Sumner LW, Yun J, Dixon RA (2013) Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol 161:1103–1116
Peel GJ, Dixon RA (2007) Detection and quantification of engineered proanthocyanidins in transgenic plants. Nat Prod Commun 2:1009–1014
Peel GJ, Pang Y, Modolo LV, Dixon RA (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59:136–149
Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32:701–712
Petrie JR, Shrestha P, Liu Q, Mansour MP, Wood CC, Zhou XR, Nichols PD, Green AG, Singh SP (2010) Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods 6:8
Prasad R, Vaid M, Katiyar SK (2012) Grape proanthocyanidins inhibit pancreatic cancer cell growth in vitro and in vivo through induction of apoptosis and by targeting the PI3K/Akt pathway. PLoS One 7:e43064
Robbins MP, Paolocci F, Hughes JW, Turchetti V, Allison G, Arcioni S, Morris P, Damiani F (2003) Sn, a maize bHLH gene, modulates anthocyanin and condensed tannin pathways in Lotus corniculatus. J Exp Bot 54:239–248
Samuel Yang S, Cheung F, Lee JJ, Ha M, Wei NE, Sze SH, Stelly DM, Thaxton P, Triplett B, Town CD (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47:761–775
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149:1028–1041
Verpoorte R (2000) Secondary metabolism. In: Alfermann AW (ed) Metabolic engineering of plant secondary metabolism. Springer, USA, pp 1–29
Wu K-M, Lu Y-H, Feng H-Q, Jiang Y-Y, Zhao J-Z (2008) Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321:1676–1678
Xiao YH, Yan Q, Ding H, Luo M, Hou L, Zhang M, Yao D, Liu HS, Li X, Zhao J, Pei Y (2014) Transcriptome and biochemical analyses revealed a detailed proanthocyanidin biosynthesis pathway in brown cotton fiber. PLoS One 9:e86344
Xie D-Y, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Xie D-Y, Sharma SB, Dixon RA (2004) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch Biochem Biophys 422:91–102
Xie D-Y, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45:895–907
Xu W, Lepiniec L, Dubos C (2014a) New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal Behav 9:e28736
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2014b) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol 202:132–144
Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49:157–169
Yu F, Barry T, Moughan P, Wilson G (1993) Condensed tannin and gossypol concentrations in cottonseed and in processed cottonseed meal. J Sci Food Agric 63:7–15
Zhao J, Dixon RA (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21:2323–2340
Zhu Y, Wang H, Peng Q, Tang Y, Xia G, Wu J, Xie D-Y (2015) Functional characterization of an anthocyanidin reductase gene from the fibers of upland cotton (Gossypium hirsutum). Planta 241:1075–1089
Acknowledgements
This study was supported by Grasslanz Technology Limited, Palmerston North, New Zealand. We thank Drs. Chenggang Liu and Ji Hyung Jun for their help with Medicago hairy root transformations. We also thank Jimmy Du from the Texas Academy of Mathematics & Science for technical assistance on this project.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, N., Roldan, M. & Dixon, R.A. Characterization of two TT2-type MYB transcription factors regulating proanthocyanidin biosynthesis in tetraploid cotton, Gossypium hirsutum . Planta 246, 323–335 (2017). https://doi.org/10.1007/s00425-017-2682-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2682-z