Abstract
Main conclusion
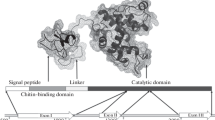
Chitinase gene from the carnivorous plant, Drosera rotundifolia , was cloned and functionally characterised.
Plant chitinases are believed to play an important role in the developmental and physiological processes and in responses to biotic and abiotic stress. In addition, there is growing evidence that carnivorous plants can use them to digest insect prey. In this study, a full-length genomic clone consisting of the 1665-bp chitinase gene (gDrChit) and adjacent promoter region of the 698 bp in length were isolated from Drosera rotundifolia L. using degenerate PCR and a genome-walking approach. The corresponding coding sequence of chitinase gene (DrChit) was obtained following RNA isolation from the leaves of aseptically grown in vitro plants, cDNA synthesis with a gene-specific primer and PCR amplification. The open reading frame of cDNA clone consisted of 978 nucleotides and encoded 325 amino acid residues. Sequence analysis indicated that DrChit belongs to the class I group of plant chitinases. Phylogenetic analysis within the Caryophyllales class I chitinases demonstrated a significant evolutionary relatedness of DrChit with clade Ib, which contains the extracellular orthologues that play a role in carnivory. Comparative expression analysis revealed that the DrChit is expressed predominantly in tentacles and is up-regulated by treatment with inducers that mimick insect prey. Enzymatic activity of rDrChit protein expressed in Escherichia coli was confirmed and purified protein exhibited a long oligomer-specific endochitinase activity on glycol-chitin and FITC-chitin. The isolation and expression profile of a chitinase gene from D. rotundifolia has not been reported so far. The obtained results support the role of specific chitinases in digestive processes in carnivorous plant species.






Similar content being viewed by others
Abbreviations
- DrChit:
-
Drosera rotundifolia chitinase
- FITC-chitin:
-
N-Fluorescein-labeled chitin
- PR:
-
Proline-rich hinge
- rDrChit:
-
Recombinant Drosera rotundifolia chitinase
References
Adrangi S, Faramarzi MA (2013) From bacteria to human: a journey into the world of chitinases. Biotechnol Adv 31:1786–1795
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bekesiova I, Nap JP, Mlynarova L (1999) Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol Biol Rep 17:269–277
Bergthorsson U, Andersson DI, Roth JR (2007) Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci USA 104:17004–17009
Bhat WW, Razdan S, Rana S, Dhar N, Wani TA, Qazi P, Vishwakarma R, Lattoo SK (2014) A phenylalanine ammonia-lyase ortholog (PkPAL1) from Picrorhiza kurroa Royle ex. Benth: molecular cloning, promoter analysis and response to biotic and abiotic elicitors. Gene 547:245–256
Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362
Bobak M, Blehova A, Kristin J, Ovecka M, Samaj J (1995) Direct plant regeneration from leaf explants of Drosera rotundifolia cultured in vitro. Plant Cell Tissue Org 43:43–49
Bonanomi A, Wiemken A, Boller T, Salzer P (2001) Local induction of a mycorrhiza-specific class III chitinase gene in cortical root cells of Medicago truncatula containing developing or mature arbuscules. Plant Biol 3:194–199
Chauhan JS, Rao A, Raghava GPS (2013) In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One 8:e67008
Clancy FG, Coffey DM (1977) Acid phosphatase and protease release by the insectivorous plant Drosera rotundifolia. Can J Bot 55:480–488
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40
Domon JM, Neutelings G, Roger D, David A, David H (2000) A basic chitinase-like protein secreted by embryogenic tissues of Pinus caribaea acts on arabinogalactan proteins extracted from the same cell lines. J Plant Physiol 156:33–39
Ďurechová D, Matušíková I, Moravčíková J, Jopčík M, Libantová J (2014) In silico analysis of chitinase promoter isolated from Drosera rotundifolia L. JMBFS 3:71–73
Ďurechová D, Matušíková I, Moravčíková J, Jopčík M, Libantová J (2015) Sequence analysis of sundew chitinase gene. JMBFS 4:4–6
Dyachok JV, Wiweger M, Kenne L, von Arnold S (2002) Endogenous Nod-factor-like signal molecules promote early somatic embryo development in Norway spruce. Plant Physiol 128:523–533
Eilenberg H, Pnini-Cohen S, Schuster S, Movtchan A, Zilberstein A (2006) Isolation and characterization of chitinase genes from pitchers of the carnivorous plant Nepenthes khasiana. J Exp Bot 57:2775–2784
Eilenberg H, Pnini-Cohen S, Rahamim Y, Sionov E, Segal E, Carmeli S, Zilberstein A (2010) Induced production of antifungal naphthoquinones in the pitchers of the carnivorous plant Nepenthes khasiana. J Exp Bot 61:911–922
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gallie DR, Chang SC (1997) Signal transduction in the carnivorous plant Sarracenia purpurea. Regulation of secretory hydrolase expression during development and in response to resources. Plant Physiol 115:1461–1471
Garcia-Casado G, Collada C, Allona I, Casado R, Pacios LF, Aragoncillo C, Gomez L (1998) Site-directed mutagenesis of active site residues in a class I endochitinase from chestnut seeds. Glycobiology 8:1021–1028
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Gomez L, Allona I, Casado R, Aragoncillo C (2002) Seed chitinases. Seed Sci Res 12:217–230
Graham LS, Sticklen MB (1994) Plant chitinases. Can J Bot 72:1057–1083
Grover A (2012) Plant chitinases: genetic diversity and physiological roles. Crit Rev Plant Sci 31:57–73
Guo XL, Bai LR, Su CQ, Shi LR, Wang DW (2013) Molecular cloning and expression of drought-induced protein 3 (DIP3) encoding a class III chitinase in upland rice. Genet Mol Res 12:6860–6870
Hatano N, Hamada T (2008) Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J Proteome Res 7:809–816
Hatano N, Hamada T (2012) Proteomic analysis of secreted protein induced by a component of prey in pitcher fluid of the carnivorous plant Nepenthes alata. J Proteomics 75:4844–4852
Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24:3439–3452
Heslop-Harrison Y (1975) Enzyme release in carnivorous plants. In: Dingle JT, Dean RT (eds) Lysosomes in biology and pathology. North-Holland, Amsterdam, pp 525–578
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587
Hunt AG (1994) Messenger-RNA 3′ end formation in plants. Annu Rev Plant Physiol 45:47–60
Ishisaki K, Arai S, Hamada T, Honda Y (2012a) Biochemical characterization of a recombinant plant class III chitinase from the pitcher of the carnivorous plant Nepenthes alata. Carbohydr Res 361:170–174
Ishisaki K, Honda Y, Taniguchi H, Hatano N, Hamada T (2012b) Heterogonous expression and characterization of a plant class IV chitinase from the pitcher of the carnivorous plant Nepenthes alata. Glycobiology 22:345–351
Islam MA, Sturrock RN, Williams HL, Ekramoddoullah AKM (2010) Identification, characterization, and expression analyses of class II and IV chitinase genes from Douglas-fir seedlings infected by Phellinus sulphurascens. Phytopathology 100:356–366
Jiang C, Huang RF, Song JL, Huang MR, Xu LA (2013) Genomewide analysis of the chitinase gene family in Populus trichocarpa. J Genet 92:121–125
Joshi CP (1987) Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res 15:9627–9640
Juniper B, Robins RJ, Joel DM (1989) The carnivorous plants. Academic Press, London, pp 1–392
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
Kirubakaran SI, Sakthivel N (2007) Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Express Purif 52:159–166
Kwon Y, Kim SH, Jung MS, Kim MS, Oh JE, Ju HW, Kim KI, Vierling E, Lee H, Hong SW (2007) Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J 49:184–193
Legrand M, Kauffmann S, Geoffroy P, Fritig B (1987) Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci USA 84:6750–6754
Li H, Greene LH (2010) Sequence and structural analysis of the chitinase insertion domain reveals two conserved motifs involved in chitin-binding. PLoS One 5:e8654
Libantova J, Kamarainen T, Moravcikova J, Matusikova I, Salaj J (2009) Detection of chitinolytic enzymes with different substrate specificity in tissues of intact sundew (Drosera rotundifolia L.). Mol Biol Rep 36:851–856
Liu ZH, Yang CP, Qi XT, Xiu LL, Wang YC (2010) Cloning, heterologous expression, and functional characterization of a chitinase gene, Lbchi32, from Limonium bicolor. Biochem Genet 48:669–679
Matusikova I, Salaj J, Moravcikova J, Mlynarova L, Nap JP, Libantova J (2005) Tentacles of in vitro-grown round-leaf sundew (Drosera rotundifolia L.) show induction of chitinase activity upon mimicking the presence of prey. Planta 222:1020–1027
Meins F, Fritig B, Linthorst HJM, Mikkelsen JD, Neuhaus J-M, Ryals J (1994) Plant chitinase genes. Plant Mol Biol Rep 12:22–28
Meszaros P, Rybansky L, Hauptvogel P, Kuna R, Libantova J, Moravcikova J, Pirselova B, Tirpakova A, Matusikova I (2013) Cultivar-specific kinetics of chitinase induction in soybean roots during exposure to arsenic. Mol Biol Rep 40:2127–2138
Michalko J, Socha P, Meszaros P, Blehova A, Libantova J, Moravcikova J, Matusikova I (2013) Glucan-rich diet is digested and taken up by the carnivorous sundew (Drosera rotundifolia L.): implication for a novel role of plant beta-1,3-glucanases. Planta 238:715–725
Mithöfer A (2011) Carnivorous pitcher plants: insights in an old topic. Phytochemistry 72:1678–1682
Neuhaus JM (1999) Plant chitinases (PR-3, PR-4, PR-8, PR-11). In: Datta SK, Mathukrishnan S (eds) Pathogenesis-related proteins in plants. CRC Press, Boca Raton, pp 77–105
Nishimura E, Kawahara M, Kodaira R, Kume M, Arai N, Nishikawa JI, Ohyama T (2013) S-like ribonuclease gene expression in carnivorous plants. Planta 238:955–967
Pan SQ, Ye XS, Kuc J (1991) A technique for detection of chitinase, beta-1,3-glucanase, and protein patterns after a single separation using polyacrylamide gel electrophoresis or isoelectrofocusing. Phytopathology 81:970–974
Passarinho PA, de Vries SC (2002) Arabidopsis chitinases: a genomic survey. In: The Arabidopsis Book 1. American Society of Plant Biologists. doi:10.1199/tab.0023
Paszota P, Escalante-Perez M, Thomsen LR, Risor MW, Dembski A, Sanglas L, Nielsen TA, Karring H, Thogersen IB, Hedrich R, Enghild JJ, Kreuzer I, Sanggaard KW (2014) Secreted major Venus flytrap chitinase enables digestion of Arthropod prey. BBA-Proteins Proteom 1844:374–383
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Razdan S, Bhat WW, Rana S, Dhar N, Lattoo SK, Dhar RS, Vishwakarma RA (2013) Molecular characterization and promoter analysis of squalene epoxidase gene from Withania somnifera (L.) Dunal. Mol Biol Rep 40:905–916
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56
Renner T, Specht CD (2012) Molecular and functional evolution of class I chitinases for plant carnivory in the Caryophyllales. Mol Biol Evol 29:2971–2985
Rottloff S, Stieber R, Maischak H, Turini FG, Heubl G, Mithöfer A (2011) Functional characterization of a class III acid endochitinase from the traps of the carnivorous pitcher plant genus, Nepenthes. J Exp Bot 62:4639–4647
Salzer P, Hebe G, Hager A (1997) Cleavage of chitinous elicitors from the ectomycorrhizal fungus Hebeloma crustuliniforme by host chitinases prevents induction of K+ and Cl− release, extracellular alkalinization and H2O2 synthesis of Picea abies cells. Planta 203:470–479
Sawant SV, Singh PK, Gupta SK, Madnala R, Tuli R (1999) Conserved nucleotide sequences in highly expressed genes in plants. J Genet 78:123–131
Schulze WX, Sanggaard KW, Kreuzer I, Knudsen AD, Bemm F, Thogersen IB, Brautigam A, Thomsen LR, Schliesky S, Dyrlund TF, Escalante-Perez M, Becker D, Schultz J, Karring H, Weber A, Hojrup P, Hedrich R, Enghild JJ (2012) The protein composition of the digestive fluid from the venus flytrap sheds light on prey digestion mechanisms. Mol Cell Proteomics 11:1306–1319
Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7:41–48
Takenaka Y, Nakano S, Tamoi M, Sakuda S, Fukamizo T (2009) Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: chitinase inhibitor allosamidin enhances stress tolerance. Biosci Biotech Biochem 73:1066–1071
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tiffin P (2004) Comparative evolutionary histories of chitinase genes in the genus Zea and family Poaceae. Genetics 167:1331–1340
Tikhonov VE, Lopez-Llorca LV, Salinas JS, Monfort E (2004) Endochitinase activity determination using N-fluorescein-labeled chitin. J Biochem Biophys Meth 60:29–38
Trudel J, Asselin A (1989) Detection of chitin deacetylase activity after polyacrylamide gel electrophoresis. Anal Biochem 189:249–253
Ubhayasekera W (2011) Structure and function of chitinases from glycoside hydrolase family 19. Polym Int 60:890–896
Vaaje-Kolstad G, Horn SJ, van Aalten DMF, Synstad B, Eijsink VGH (2005) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem 280:28492–28497
Veluthakkal R, Dasgupta MG (2012) Isolation and characterization of pathogen defence-related class I chitinase from the actinorhizal tree Casuarina equisetifolia. Forest Pathol 42:467–480
Verburg JG, Rangwala SH, Samac DA, Luckow VA, Huynh QK (1993) Examination of the role of tyrosine-174 in the catalytic mechanism of the Arabidopsis thaliana chitinase: comparison of variant chitinases generated by site-directed mutagenesis and expressed in insect cells using baculovirus vectors. Arch Biochem Biophys 300:223–230
Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J (2009) SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res 37:D380–D386
Wu XF, Wang CL, Xie EB, Gao Y, Fan YL, Liu PQ, Zhao KJ (2009) Molecular cloning and characterization of the promoter for the multiple stress-inducible gene BjCHI1 from Brassica juncea. Planta 229:1231–1242
Zhao JW, Wang JL, An LL, Doerge RW, Chen ZJ, Grau CR, Meng JL, Osborn TC (2007) Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227:13–24
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Acknowledgments
This work was supported by a grant from the Slovak Grant Agency VEGA No. 2/0090/14 and MVTS COST Action FA1208.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jopcik, M., Moravcikova, J., Matusikova, I. et al. Structural and functional characterisation of a class I endochitinase of the carnivorous sundew (Drosera rotundifolia L.). Planta 245, 313–327 (2017). https://doi.org/10.1007/s00425-016-2608-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2608-1