Abstract
Main conclusion
The present study documented the predominant role of WRKY transcription factor in controlling genes of different pathways related to fibre formation in jute and could be a candidate gene for the improvement of jute fiber.
Abstract
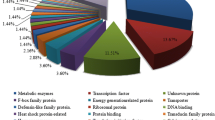
Understanding of molecular mechanism associated with bast fibre development is of immense significance to achieve desired improvement in jute (Corchorus sp.). Therefore, suppression subtractive hybridization was successfully applied to identify genes involved in fibre developmental process in jute. The subtracted library of normal Corchorus capsularis as tester with respect to its fibre-deficient mutant as driver resulted in 2,685 expressed sequence tags which were assumed to represent the differentially expressed genes between two genotypes. The identified expressed sequence tags were assembled and clustered into 225 contigs and 231 singletons. Among these 456 unigenes, 377 were classified into 15 different functional categories while others were of unknown functional category. Reverse Northern analysis of the unigenes showed distinct variation in hybridization intensity of 11 transcripts between two genotypes tested. The findings were also documented by Northern and real-time PCR analysis. Varied expression level of these transcripts suggested their crucial involvement in fibre development in this species. Among these transcripts, WRKY transcription factor was documented to be a most important transcript which was in agreement with its known role in other plant species in possible regulation related to cell wall biosynthesis, expansion and lignification. This report constitutes first systematic analysis of genes involved in fibre development process in jute. It may be suggested that the information generated in this study would be useful for genetic improvement of fibre traits in this plant species.





Similar content being viewed by others
Abbreviations
- AGP:
-
Arabinogalactan protein
- CAD:
-
Cinnamyl alcohol dehydrogenase
- CCR:
-
Cinnamoyl-CoA reductase
- C4H:
-
Cinnamate-4-hydroxylase
- CHS:
-
Chalcone synthase
- 4CL:
-
4-Coumarate CoA:ligase
- COMT:
-
Caffeic acid 3-O-methyltransferase
- Das:
-
Days after sowing
- EST:
-
Expressed sequence tags
- FF:
-
Fibre forming
- F5H:
-
Ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydoxylase
- GRP:
-
Glycine rich protein
- HCT:
-
Hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase
- HD-Zip:
-
Homeodomain leucine zipper protein
- PAL:
-
Phenylalanine ammonia lyase
- PEL:
-
Pectate lyase
- PME:
-
Pectin methylesterase
- SSH:
-
Suppression subtractive hybridization
- XET:
-
Xyloglucan endotransglycosylase
- ZFP:
-
Zinc finger protein
References
Abid G, Muhoviski Y, Jacquemin J-M, Mingeot D, Sassi K, Toussaint A, Baudoin JP (2011) Changes in DNA-methylation during zygotic embryogenesis in interspecific hybrids of beans (Phaseolus sp.). Plant Cell Tiss Organ Cult 105:383–393
Bausher M, Shatters R, Chaparro J, Dang P, Hunter W, Niedz R (2003) An expressed sequence tag (EST) set from Citrus sinensis L. Osbeck whole seedlings and the implications of further perennial source investigations. Plant Sci 165:415–422
Bedon F, Grima-Pettenati J, Mackay J (2007) Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol 7:17–34
Blake AW, Marcus SE, Copeland JE, Blackburn RS, Knox JP (2008) In situ analysis of cell wall polymers associated with phloem fibre cells in stems of hemp, Cannabis sativa L. Planta 228:1–13
Bolton JJ, Soliman KM, Wilkins TA, Jenkins JN (2009) Aberrant expression of critical genes during secondary cell wall biogenesis in a cotton mutant, Ligon Lintless-1 (Li-1). Comp Funct Genomics 2009:1–8
Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14:3073–3088
Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407:321–326
Côté CL, Boileau F, Roy V, Ouellet M, Levasseur C, Morency MJ, Cooke JEK, Séguin A, Côté JJM (2010) Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol 10:273
Day A, Addi M, Kim W, David H, Bert F, Mesnage P, Rolando C, Chabbert B, Neutelings G, Hawkins S (2005) ESTs from the fibre-bearing stem tissues of flax (Linum usitatissimum L.): expression analyses of sequences related to cell wall development. Plant biol 7:23–32
Demura T, Fukuda H (2007) Transcriptional regulation in wood formation. Trends Plant Sci 12:64–70
Dharmawardhana P, Brunner AM, Strauss SH (2010) Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genom 11:150–169
Dou L, Zhang X, Pang C, Song M, Wei H, Fan S, Yu S (2014) Genome-wide analysis of the WRKY gene family in cotton. Mol Genet Genomics. doi:10.1007/s00438-014-0872-y
Du J, Miura E, Robischon M, Martinez C, Groover A (2011) The Populus class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS One 6:e17458
Elhiti M, Stasolla C (2009) Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav 4:86–88
Gea G, Kjell S, Jean-François H (2013) Integrated -Omics: a powerful approach to understanding the heterogeneous lignification of fibre crops. Int J Mol Sci 14:10958–10978
Ghosh T (1983) Handbook on jute. FAO plant production and protection. Paper No. 51. Rome: FAO
Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, Trossat-Magnin C, Delrot S, Lauvergeat V (2010) The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 72:215–234
Harris JC, Hrmova M, Lopato S, Langridge P (2011) Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol 190:823–837
Herbers K, Lorences EP, Barrachina C, Sonnewald U (2001) Functional characterization of Nicotiana tabacum xyloglucan endotransglycosylase (Nt XET-1): generation of transgenic tobacco plants and changes in cell wall xyloglucan. Planta 212:279–287
Hisano H, Nandakumar R, Wang ZY (2009) Genetic modification of lignin biosynthesis for improved biofuel production. In Vitro Cell Dev Biol-Plant 45:306–313
Hofer J, Turner L, Moreau C, Ambrose M, Isaac P, Butcher S, Weller J, Dupin A, Dalmais M, Le Signor C, Bendahmane A, Ellis N (2009) Tendril-less regulates tendril formation in pea leaves. Plant Cell 21:420–428
Hsing YI, Cher CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, Huang WL, Ko SS, Chen S, Chen JL, Chung CI, Lin YC, Hour AL, Wang YW, Chang YC, Tsai MW, Lin YS, Chen YC, Yen HM, Li CP, Wey CK, Tseng CS, Lai MH, Huang SC, Chen LJ, Yu SM (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63:351–364
Jensen WA (1962) Botanical histochemistry—principles and practice. Freeman and Company, University of California, Berkeley
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635
Lacoux J, Klein D, Domon JM, Burel C, Lamblin F, Alexandre F, Sihachakr D, Roger D, Balange AP, David A, Morvan C, Lainé E (2003) Antisense transgenesis of Linum usitatissimum with a pectin methylesterase cDNA. Plant Physiol Biochem 41:241–249
Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD (2010) Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232:1191–1205
Leida C, Romeu JF, García-Brunton J, Ríos G, Badenes ML (2012) Gene expression analysis of chilling requirements for flower bud break in peach. Plant Breed 131:329–334
Li X, Weng JK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54:569–581
Li Y, Liu D, Tu L, Zhang X, Wang L, Zhu L, Tan J, Deng F (2010) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep 29:193–202
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408
Long SH, Deng X, Wang YF, Li X, Qiao RQ, Qiu CS, Guo Y, Hao DM, Jia WQ, Chen XB (2012) Analysis of 2,297 expressed sequence tags (ESTs) from a cDNA library of flax (Linum ustitatissimum L.) bark tissue. Mol Biol Rep 39:6289–6296
Ma Y, Hong Z, Wu Q, Yao X (2011) Transcriptome subtractive hybridization and its reliability validated by an E. coli model. Biosci Method 2:38–42
Mangeon A, Junqueira RM, Sachetto-Martins G (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5:99–104
Meredith R (1956) Mechanical properties of textile fibres, vol Chapters VI and VII. Interscience Publisher, New York
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
Motamayor JC, Mockaitis K, Schmutz J, Haiminen N, Livingstone D III, Cornejo O, Findley SD, Zheng P, Utro F, Royaert S, Saski C, Jenkins J, Podicheti R, Zhao M, Scheffler BE, Stack JC, Feltus FA, Mustiga GM, Amores F, Phillips W, Marelli JP, May GD, Shapiro H, Ma J, Bustamante CD, Schnell RJ, Main D, Gilbert D, Parida L, Kuhn DN (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol 14:1–24
Nishikubo N, Takahashi J, Roos AA, Derba-Maceluch M, Piens K, Brumer H, Teri TT, Stalbrand H, Mellerowicz EJ (2011) Xyloglucan endo transglycosylase mediated xyloglucan rearrangements in developing wood of hybrid Aspen. Plant Physiol 155:399–413
Orfila C, Huisman MMH, Willats WGT, Van Alebeek GJWM, Schols HA, Seymour GB, Knox JP (2001) Altered middle lamella homogalacturonan and disrupted deposition of (1 → 5)-α-l-arabinan in the pericarp of cnr, a ripening mutant of tomato. Plant Physiol 126:210–221
Pelloux J, Rustérucci C, Mellerowicz EJ (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12:267–277
Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23:2209–2224
Pritchard J (1994) The control of cell expansion in roots. New Phytol 127:3–26
Rastogi S, Dwivedi UN (2008) Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Sci 174:264–277
Ringlia C, Keller B, Ryse U (2001) Glycine-rich proteins as structural components of plant cell walls. Cell Mol Life Sci 58:1430–1441
Roy MM (1953) Mechanical properties of jute. The study of chemically treated fibres. J Tex Inst Trans 44:44–52
Sadhukhan S (2007) Characterization of jute fibre and generation of developmental stage specific expressed sequence tags in relation to fibre formation. PhD Thesis, Indian Institute of Technology, Kharagpur, West Bengal
Sahu BB, Shaw BP (2009) Isolation, identification and expression analysis of salt-induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biol 9:69
Samanta P, Sadhukhan S, Das S, Joshi A, Sen SK, Basu A (2011) Isolation of RNA from field-grown jute (Corchorus capsularis) plant in different developmental stages for effective downstream molecular analysis. Mol Biotechnol 49:109–115
Sanchez P, Nehlin L, Greb T (2012) From thin to thick: major transitions during stem development. Trends Plant Sci 17:113–121
Sengupta G, Palit P (2004) Characterization of a lignified secondary phloem fibre-deficient mutant of jute (Corchorus capsularis). Ann Bot 93:211–220
Siedlecka A, Wiklund S, Pe´ronne M-A, Micheli F, Les´niewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ (2008) Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of populus. Plant Physiol 146:554–565
Sonbol F-M, Fornalé S, Capellades M, Encina A, Touriño S, Torres J-L, Rovira P, Ruel K, Puigdomènech P, Rigau J, Caparrós-Ruiz D (2009) The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol Biol 70:283–296
Stephens J, Halpin C (2007) Lignin manipulation for fibre improvement. In: Ranalli P (ed) Improvement of crop plants for industrial end uses, chapter 5. Springer, Netherlands, pp 129–153
Sui S, Luo J, Ma J, Zhu Q, Lei X, Li M (2011) Generation and analysis of expressed sequence tags from Chimonanthus praecox (Wintersweet) flowers for discovering stress-responsive and floral development-related genes. Comp Func Genomics. doi:10.1155/2012/134596
Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10:135–154
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Van den Broeck HC, Maliepaard C, Ebskamp MJM, Toonen MAJ, Koops AJ (2008) Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.). Plant Sci 174:205–220
Wang HZ, Dixon RA (2011) On–off switches for secondary cell wall biosynthesis. Mol Plant 5:297–303
Wang H, Guo Y, Lv F, Zhu H, Wu S, Jiang Y, Li F, Zhou B, Guo W, Zhang T (2010) The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol Biol 72:397–406
Wang H, Zhao Q, Chen F, Wang M, Dixon RA (2011) NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 68:1104–1114
Wei KF, Chen J, Chen YF, Wu LJ, Xie DX (2012) Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res 19:153–164
Yadun SL (2010) Plant fibers: initiation, growth, model plants, and open questions. Russ J Plant Physiol 57:305–315
Yang J, Showalter AM (2007) Expression and localization of AtAGP18, a lysine-rich arabinogalactan-protein in Arabidopsis. Planta 226:169–179
Zhang JZ, Li ZM, Liu L, Mei L, Yao JL, Hu CG (2008) Identification of early-flower-related ESTs in an early-flowering mutant of trifoliate orange (Poncirus trifoliata L. Raf.) by suppression subtractive hybridization and macroarray analysis. Tree Physiol 28:1449–1457
Zhong R, Ye ZH (1999) IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis encodes a homeodomain-leucine zipper protein. Plant Cell 11:2139–2152
Acknowledgments
Financial assistance in the form the grant support to this laboratory from the Department of Biotechnology, Government of India is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Samanta, P., Sadhukhan, S. & Basu, A. Identification of differentially expressed transcripts associated with bast fibre development in Corchorus capsularis by suppression subtractive hybridization. Planta 241, 371–385 (2015). https://doi.org/10.1007/s00425-014-2187-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2187-y