Abstract
Background
Technical improvements in magnetic resonance imaging (MRI) acquisition, such as higher field strength and optimized sequences, lead to better multiple sclerosis (MS) lesion detection and characterization. Multiplication of 3D-FLAIR with 3D-T2 sequences (FLAIR2) results in isovoxel images with increased contrast-to-noise ratio, increased white–gray-matter contrast, and improved MS lesion visualization without increasing MRI acquisition time. The current study aims to assess the potential of 3D-FLAIR2 in detecting cortical/leucocortical (LC), juxtacortical (JC), and white matter (WM) lesions.
Objective
To compare lesion detection of 3D-FLAIR2 with state-of-the-art 3D-T2-FLAIR and 3D-T2-weighted images.
Methods
We retrospectively analyzed MRI scans of thirteen MS patients, showing previously noted high cortical lesion load. Scans were acquired using a 3 T MRI scanner. WM, JC, and LC lesions were manually labeled and manually counted after randomization of 3D-T2, 3D-FLAIR, and 3D-FLAIR2 scans using the ITK-SNAP tool.
Results
LC lesion visibility was significantly improved by 3D-FLAIR2 in comparison to 3D-FLAIR (4 vs 1; p = 0.018) and 3D-T2 (4 vs 1; p = 0.007). Comparing LC lesion detection in 3D-FLAIR2 vs. 3D-FLAIR, 3D-FLAIR2 detected on average 3.2 more cortical lesions (95% CI − 9.1 to 2.8). Comparing against 3D-T2, 3D-FLAIR2 detected on average 3.7 more LC lesions (95% CI 3.3–10.7).
Conclusions
3D-FLAIR2 is an easily applicable time-sparing MR post-processing method to improve cortical lesion detection. Larger sampled studies are warranted to validate the sensitivity and specificity of 3D-FLAIR2.
Similar content being viewed by others
Introduction
Multiple sclerosis (MS) is a chronic inflammatory demyelinating disease of the central nervous system (CNS) in which magnetic resonance imaging (MRI) is an invaluable diagnostic tool in establishing the diagnosis [22]. Improved lesion detection either by increasing field strength or improving acquisition and post-processing techniques possibly beneficially impacts time to diagnosis and disease monitoring [2, 25, 26].
While white matter (WM) lesions have long been the main focus of research in MS, cortical lesions (CL) are more and more recognized as playing an important role [12]. In the recent update of the diagnostic imaging criteria, the detection of cortical und juxtacortical (JC) lesions received additional weight [22]. In contrast to WM lesions, which are easily identified using standard MRI techniques [8], less than 25% of histopathologically confirmed CL are detectable by conventional (FLAIR and T2) clinical MR imaging [4]. Furthermore, both white and gray matter lesions have been associated with disability accumulation and potential outcome parameters in clinical trials [7, 10]. Therefore, there is a specific demand for high sensitivity in lesion detection of CL and JC lesions by means of MRI.
Current guidelines for standardized brain and spinal cord MRI for the diagnosis and follow-up of MS include 3D-T2-FLAIR and 3D-T2-weighted images [13, 23]. It was recently shown that the combination of 3D-T2 weighted images with 3D-FLAIR, referred to as 3D-FLAIR2, leads to a better contrast-to-noise ratio and white–gray-matter contrast, while still suppressing CSF signals and thereby to improved lesion visualization without the need for additional scan time [27].
In this monocentric study, we aimed to test the practicability of retrospectively computed 3D-FLAIR2 images of routinely acquired MRIs. We tried to prove or reject the hypothesis that 3D-FLAIR2 shows a higher sensitivity compared to 3D-T2-FLAIR or 3D-T2-weighted images alone in detecting cortical, LC, JC, and WM lesions in MS patients.
Materials and methods
Ethics
The study was approved by the ethics committee of the Medical University Vienna (ethical approval number: EK1464/2017).
Patients and definitions
This is a retrospective analysis of patients recruited from the Department of Neurology, Medical University of Vienna between 2017 and 2018. The first thirteen MS patients from the dataset from the Vienna MS database (VMSD), with a high lesion load noted on previously addressed MRIs, were included in this study. All patients fulfilled the McDonald MS criteria [22].
Image acquisition
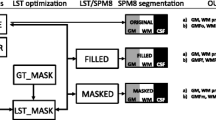
MRI scans were acquired using a 3 T Achieva Philips Healthcare MRI scanner using an 8-channel SENSE head coil. 3D-FLAIR: voxel size = 0.67 mm × 0.76 mm × 1.34 mm; TR = 4800 ms; TE = 415 ms; TI = 1650 ms; Fat suppression: 3D-T2 = TR – 2500 ms, TE = 314 ms, acquired voxel size = 0.67 mm × 0.76 mm × 1.34 mm, reconstructed voxel size: 0.65 mm × 0.65 mm × 0.65 mm; SPIR; acquired voxel size = 0.67 mm × 0.76 mm × 1.34 mm; reconstructed voxel size = 0.65 mm × 0.65 mm × 0.65 mm; post sequences were reconstructed in three orthogonal planes.
Data processing was performed with Advanced Normalization Tools (ANTs v2.2.0) and FSL (v6.0). Both, T2 and FLAIR volumes were bias field corrected using N4 [24], and subsequently, image intensities were normalized. FLAIR images were co-registered and resampled to the T2 volume space using FLIRT [14, 15] (12 degrees of freedom, mutual information, sinc interpolation). Finally, the 3-DFLAIR2 image was obtained by multiplication of the aligned FLAIR and T2 volumes.
Image analysis
Image analysis includes total white matter lesion counts of WM, JC, and cortical/LC lesions. Lesions were manually labeled by two trained raters (TZ, PR) with 5 and 10 years of experience in MS imaging, randomly and blinded in 3D-T2, 3D-FLAIR, and 3D-FLAIR2 with the ITK-SNAP tool [28]. In the case of disagreement, a senior neuroradiological rater with more than 15 years of experience in MS imaging was consulted (GK) to reach consensus. Lesions were defined according to previous literature [9].
Statistics
Statistical analysis was performed using IBM SPSS 20.0.0 (SPSS Inc, Chicago, IL, USA) and R studio (Version 1.2.5033, RStudio, Inc.). The power analysis was based on previous data on WM/CL lesion subtype-specific frequencies in different stages of the disease in individuals with MS [12] (p value 0.05, power 80%) [20].
Normal distribution was assessed and rejected for lesion count variables with Shapiro–Wilk’s method, which are provided with median value and interquartile range. Differences between two groups were assessed with the Mann–Whitney U test. Bland–Altman plots were calculated to quantify the amount of agreement in lesion counts derived from different sequences (3D-FLAIR2, 3D-FLAIR, and 3D-T2) and different raters (TZ vs. PR TZ vs. TZ) for WM, JC, and LC lesions. The mean difference and the limits of agreement, which reflect the 95% level as described by Bland and Altman [3] are provided with their 95% CI.
Inter- and intra-rater variability was assessed with intraclass correlation coefficient for all sequences using a two-way random-effects model with absolute agreement [17]. Intra-rater variability was assessed by two independent lesion counts by rater TZ (TZ1 vs. TZ2), while inter-rater variability was assessed by calculating lesion assessment of (TZ1 vs PR1). Significance was set at a two-sided p value of 0.05.
Data availability
Anonymized data not published in the article can be made available upon reasonable request from a qualified investigator after approval from the ethics review board of the Medical University of Vienna.
Results
Patient characteristics
13 patients with a confirmed diagnosis of MS [3 males and 10 females, mean age 37.3 years ± 13.9 (SD)] were included. The median EDSS score was 2.5; 4.5 (IQR). The mean disease duration was 6.3 years ± 6.0 (SD). Nine patients were classified as having a relapsing remittent disease course, three patients presented a secondary progressive and one patient with a primary progressive disease course.
Lesion detection
A total of 1067 3D-FLAIR2, 809 3D-FLAIR, and 577 3D-T2-weighted lesions were detected in these 13 patients.
WM lesions were more common on 3D-FLAIR2 sequences compared to 3D-FLAIR (median 52 vs 40, p = 0.37) and 3D-T2-weighted scans (median 52 vs 34, p = 0.077) (Figs. 1, 2).
3D-FLAIR2 lesion visualization compared to 3D-T2 and 3D-FLAIR. A-C: Depiction of a cortical/leucocortical lesion on axial 3D-FlAIR2 (A), 3D-FLAIR (B), 3D-T2 (C) MRI images. D–F Presentation of a large white matter lesion with an adjacent cortical/leucocortical lesion. G–I Juxtacortical lesion involving the right frontal superior gyrus; J–L Juxtacortical U-fiber lesion on sagittal view. M–O Temporal cortical/leucocortical lesion on sagittal view
Overall, 70 JC were counted. In 3D-FLAIR2 images more JC lesions were counted compared to 3D-FLAIR (median 2 vs 1; p = 0.54) and 3D-T2-weighted images (median 2 vs 0; p = 0.11), however not reaching statistically significance.
LC lesion visibility was significantly improved by 3D-FLAIR2 in comparison to 3D-FLAIR (median 4 vs 1; p = 0.018) and 3D-T2 (median: 4 vs 1; p = 0.007). Comparing LC lesion detection in 3D-FLAIR2 vs 3D-FLAIR, 3D-FLAIR2 detected on average 3.2 more cortical lesions (95% CI − 9.1 to 2.8) per patient. Comparing against 3D-T2, 3D-FLAIR2 detected on average 3.7 more LC lesions (95% CI 3.3–10.7) (Fig. 3).
Difference in the numbers of assessed lesions. Bland–Altman plots comparing lesion counts derived from 3D-FLAIR2, 3D-FLAIR and 3D-T2 in each patient and each location. For white matter lesion (WML), juxtacortical lesion (JCL) and leucocortical lesion (LCL), the difference between lesion counts derived from the different sequences (3D-FLAIR2, 3D-FLAIR and T2), is plotted relative to their mean for each patient with black dots. The black dashed line provides the mean difference with the corresponding 95% CI in blue. The limits of agreement are provided with their 95% CI, the upper bound in green, the lower bound in red
We further calculated the interclass correlation coefficients (ICC) to quantify the amount of correlation adjusted for random effects between lesion counts obtained from different sequences (Fig. 4). The highest ICC was calculated for WM lesion followed by those from JC and LC lesion metrics.
JC lesions measured on 3D-T2 vs 3D-FLAIR2 (0.30 ICC, 95% CI − 0.27 to 0.72) or 3D-FLAIR (0.16 ICC, 95% CI − 0.40 to 0.64) and LC lesions in 3D-T2 vs 3D-FLAIR2 (0.41 ICC, 95% CI − 0.15 to 0.78) displayed a 95% CI involving zero.
To determine the reproducibility for 3D-FLAIR2 intra-rater and intra-correlation coefficients were calculated 0.90 (WM), 0.88 (JC) and 0.86 (LC). Interrater intra-correlation for 3D-FLAIR2 were 0.91 (WM), 0.78 (JC) and 0.75 (LC) (Suppl. Figure 1/Suppl Table 1).
Discussion
Detection of lesions by MRI is an integral component of both diagnosis and disease monitoring in MS [22, 23]. Here, we assessed the added diagnostic value of a voxel-wise multiplication of 3D-T2 weighted images with 3D-FLAIR images, resulting in 3D-FLAIR2 compared to the acquired initially standard 3D sequences alone. We demonstrated that 3D-FLAIR2 increases the detection rate of LC lesions compared with state-of-the-art T2 and FLAIR 3D sequences.
The potential benefit of 3D-FLAIR2 was previously shown by demonstrating a higher contrast-to-noise ratio for WM and GM lesions in comparison to FLAIR or T2 images [27]. It was proposed that this approach produces a similar contrast like double inversion recovery (DIR), however, with improved image quality and less acquisition time. While FLAIR2 was first proposed with 3D scans, it can also be used with 2D-FLAIR and 2D-T2 scans [18]
In line with these data, we provide evidence that 3D-FLAIR2 outperformed state-of-the-art 3D-FLAIR and 3D-T2-weighted sequences in lesion visualization of cortical lesions without the need for additional image acquisition time. 3D-FLAIR2 is a sensitive and radiologically feasible tool in clinical routine and clinical studies for MS lesion assessment at not cost of imaging time. As 3D-FLAIR2 may also improve automatic lesion segmentation, it could easily be implemented in future automatic MS lesion detection algorithms [18]. Automatized segmentation and Artificial intelligence (AI) of MRI images have great potential in monitoring disease activity in demyelinating diseases of the central nervous system and guiding diagnostic pathways [1, 16, 19], 3D-FLAIR2 may further improve the quality of diagnosing and monitoring these patients non-invasively. Likewise, it will also increase human capabilities in lesion detection and potentially help in the supervision of machine learning.
As WM lesion load only in part explains clinical disease progression, conversion and cognitive decline, cortical lesions increasingly become a focus of research [5]. Despite advances in imaging, even under optimal conditions, a maximum of 25% of the actual dimension can be visualized in histopathology correlation studies [4]. Improving cortical lesion detection has the potential to improve the prediction of subsequent disease evolution and therapeutic response and as well as to improve fulfilling the criterion of dissemination in space [6, 22]. Here, we could show that 3D-FLAIR2 can enhance the detection of cortical/leucocortical lesions compared to standard routine sequences and display excellent inter and intra-rater variability (Figs. 3,4).
It should be noted that our sample consists of preselected MS patients with previously noted high cortical lesion load. Further limitations of this study include its retrospective nature. We used 3D images as reference images for comparison since they outperform lesion detection compared to 2D images [11, 21]. We could not compare our findings to PSIR and DIR images, as these are not part of the used MS MRI protocols. Therefore, future prospective studies should include a larger number of subjects, potential histopathological correlations, and direct comparison with DIR/PSIR to determine the value of 3D-FLAIR2 in improving disease monitoring and MS diagnosis.
In summary, we show that combining 3D-T2 and 3D-FLAIR sequence data to create 3D-FLAIR2 is a feasible and easily applicable strategy to specifically improve cortical/leucocortical lesion detection in MS.
Change history
26 October 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00415-021-10855-5
References
Afzal HR, Luo S, Ramadan S, Lechner-Scott J (2020) The emerging role of artificial intelligence in multiple sclerosis imaging. Mult Scler J. https://doi.org/10.1177/1352458520966298
Bink A, Schmitt M, Gaa J, Mugler JP, Lanfermann H, Zanella FE (2006) Detection of lesions in multiple sclerosis by 2D FLAIR and single-slab 3D FLAIR sequences at 3.0 T: initial results. Eur Radiol 16:1104–1110
Bland JM, Altman D (1986) Statistical methods for assessing agreement between two methods of clinical measurement. The lancet 327:307–310
Bouman PM, Steenwijk MD, Pouwels PJ, Schoonheim MM, Barkhof F, Jonkman LE, Geurts JJ (2020) Histopathology-validated recommendations for cortical lesion imaging in multiple sclerosis. Brain 143(10):2988–2997
Calabrese M, Agosta F, Rinaldi F, Mattisi I, Grossi P, Favaretto A, Atzori M, Bernardi V, Barachino L, Rinaldi L (2009) Cortical lesions and atrophy associated with cognitive impairment in relapsing-remitting multiple sclerosis. Arch Neurol 66:1144–1150
Calabrese M, Filippi M, Gallo P (2010) Cortical lesions in multiple sclerosis. Nat Rev Neurol 6:438
Filippi M, Agosta F (2010) Imaging biomarkers in multiple sclerosis. J Magn Reson Imaging 31:770–788
Filippi M, Brück W, Chard D, Fazekas F, Geurts JJ, Enzinger C, Hametner S, Kuhlmann T, Preziosa P, Rovira À (2019) Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol 18:198–210
Filippi M, Preziosa P, Banwell BL, Barkhof F, Ciccarelli O, De Stefano N, Geurts JJ, Paul F, Reich DS, Toosy AT (2019) Assessment of lesions on magnetic resonance imaging in multiple sclerosis: practical guidelines. Brain 142:1858–1875
Goldman MD, Motl RW, Rudick RA (2010) Possible clinical outcome measures for clinical trials in patients with multiple sclerosis. Ther Adv Neurol Disord 3:229–239
Gramsch C, Nensa F, Kastrup O, Maderwald S, Deuschl C, Ringelstein A, Schelhorn J, Forsting M, Schlamann M (2015) Diagnostic value of 3D fluid attenuated inversion recovery sequence in multiple sclerosis. Acta Radiol 56:622–627
Haider L, Prados F, Chung K, Goodkin O, Kanber B, Sudre C, Yiannakas M, Samson RS, Mangesius S, Thompson AJ (2021) Cortical involvement determines impairment 30 years after a clinically isolated syndrome. Brain 144(5):1384–1395
Hu XY, Rajendran L, Lapointe E, Tam R, Li D, Traboulsee A, Rauscher A (2019) Three-dimensional MRI sequences in MS diagnosis and research. Mult Scler J 25:1700–1709
Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841
Jenkinson M, Smith S (2001) A global optimisation method for robust affine registration of brain images. Med Image Anal 5:143–156
Kanber B, Nachev P, Barkhof F, Calvi A, Cardoso J, Cortese R, Prados F, Sudre CH, Tur C, Ourselin S (2019) High-dimensional detection of imaging response to treatment in multiple sclerosis. NPJ Digit Med 2:1–10
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163
Le M, Tang L, Hernández-Torres E, Jarrett M, Brosch T, Metz L, Li D, Traboulsee A, Tam R, Rauscher A (2019) FLAIR2 improves LesionTOADS automatic segmentation of multiple sclerosis lesions in non-homogenized, multi-center, 2D clinical magnetic resonance images. Neuroimage 23:101918
Lladó X, Oliver A, Cabezas M, Freixenet J, Vilanova JC, Quiles A, Valls L, Ramió-Torrentà L, Rovira À (2012) Segmentation of multiple sclerosis lesions in brain MRI: a review of automated approaches. Inf Sci 186:164–185
Peterson SJ, Foley S (2021) Clinician’s guide to understanding effect size, α level, power, and sample size. Nutr Clin Pract 36(3):598–605
Tan I, Pouwels P, van Schijndel R, Adèr H, Manoliu R, Barkhof F (2002) Isotropic 3D fast FLAIR imaging of the brain in multiple sclerosis patients: initial experience. Eur Radiol 12:559–567
Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, Correale J, Fazekas F, Filippi M, Freedman MS, Fujihara K, Galetta SL, Hartung HP, Kappos L, Lublin FD, Marrie RA, Miller AE, Miller DH, Montalban X, Mowry EM, Sorensen PS, Tintoré M, Traboulsee AL, Trojano M, Uitdehaag BMJ, Vukusic S, Waubant E, Weinshenker BG, Reingold SC, Cohen JA (2018) Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 17:162–173
Traboulsee A, Simon J, Stone L, Fisher E, Jones D, Malhotra A, Newsome S, Oh J, Reich D, Richert N (2016) Revised recommendations of the consortium of MS centers task force for a standardized MRI protocol and clinical guidelines for the diagnosis and follow-up of multiple sclerosis. Am J Neuroradiol 37:394–401
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Wattjes MP, Harzheim M, Lutterbey GG, Hojati F, Simon B, Schmidt S, Schild HH, Barkhof F (2008) Does high field MRI allow an earlier diagnosis of multiple sclerosis? J Neurol 255:1159–1163
Wattjes MP, Lutterbey GG, Harzheim M, Gieseke J, Träber F, Klotz L, Klockgether T, Schild HH (2006) Higher sensitivity in the detection of inflammatory brain lesions in patients with clinically isolated syndromes suggestive of multiple sclerosis using high field MRI: an intraindividual comparison of 1.5 T with 3.0 T. Eur Radiol 16:2067–2073
Wiggermann V, Hernandez-Torres E, Traboulsee A, Li D, Rauscher A (2016) FLAIR2: a combination of FLAIR and T2 for improved MS lesion detection. Am J Neuroradiol 37:259–265
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128
Funding
Open access funding provided by Medical University of Vienna. There was no funding to this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Zrzavy Tobias has no conflict of interest relevant to this study. Wielandner Alice has no conflict of interest relevant to this study. Haider Lukas has no conflict of interest relevant to this study. Bartsch Sophie has no conflict of interest relevant to this study. Leutmezer Fritz has no conflict of interest relevant to this study. Berger Thomas has nothing to disclose for this study. Nenning Karl-Heinz has no conflict of interest relevant to this study. Rauscher Alexander has no conflict of interest to declare. Rommer Paulus has received honoraries for lectures/consultancy from AbbVie, Alexion, Almirall, Biogen, Merck, Novartis, Sandoz, Teva, has received research grants from Biogen, Merck, Roche. None resulted in a conflict of interest with regard to the submitted manuscript. Kasprian Gregor has no conflict of interest relevant to this study.
Ethical standards
The study was approved by the ethics committee of the Medical University of Vienna and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Additional information
The original online version of this article was revised: the given and family names were incorrectly structured.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zrzavy, T., Wielandner, A., Haider, L. et al. FLAIR2 post-processing: improving MS lesion detection in standard MS imaging protocols. J Neurol 269, 461–467 (2022). https://doi.org/10.1007/s00415-021-10833-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-021-10833-x