Abstract
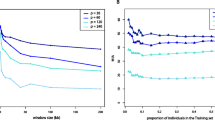
An enlarged mtDNA database (n=549) for the Portuguese population, comprising HVRI and HVRII regions is reported. This database was used to test the effect of sample size on the estimation of relevant parameters such as haplotype diversity, number of different haplotypes, nucleotide diversity and number of polymorphic positions. Simulations were performed generating sets of random subsamples of variable sizes (n=50, 100, 200, 300 and 400). The results show that while haplotype and nucleotide diversities do not vary significantly with sample size, the numbers of haplotypes and polymorphic positions rise continuously inside the tested interval. These trends are interpretable by the evolution of the proportions of sequences that are found once or twice, which drop dramatically as sample size increases, with the corresponding rise in the frequency of those encountered 3 times or more. The generated data were also used to extrapolate saturation curves for the referred parameters. When considering for instance the number of haplotypes, it is shown that a sample size of 1,000 individuals is required for practical saturation (defined as the point where a sample size increase of 100 individuals corresponds to an increment in the diversity measure below 5%). For HVRII the same level is reached at n=900 and n=1,300 is needed when both regions are analysed simultaneously. Consequently, we can infer that currently used sample sizes are still rather inadequate for both anthropological and forensic purposes.




Similar content being viewed by others
References
Anderson S, Bankier AT, Barrell BG et al.(1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Arnason E (2003) Genetic heterogeneity of Icelanders. Ann Hum Genet 67:5–16
Bendall KE, Sykes BC (1995) Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet 57:248–256
Forster L, Forster P, Lutz-Bonengel S, Willkomm H, Brinkmann B (2002) Natural radioactivity and human mitochondrial DNA mutations. Proc Natl Acad Sci U S A 99:13950–13954
Helgason A, Sigurðardóttir S, Gulcher J, Stefánsson K, Ward R (2000) Sampling saturation and the European mtDNA pool: implications for detecting genetic relationships among populations. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research, Cambridge, pp 285–294
Pereira L, Prata MJ, Amorim A (2000) mtDNA diversity in Portugal: not a genetic edge of European variation. Ann Hum Genet 64:491–506
Pfeiffer H, Brinkmann B, Hühne J et al. (1999) Expanding the forensic German mitochondrial DNA control region database: genetic diversity as a function of sample size and microgeography. Int J Legal Med 112:291–298
Pfeiffer H, Forster P, Ortmann C, Brinkmann B (2001) The results of an mtDNA study of 1200 inhabitants of a German village in comparison to other Caucasian databases and its relevance for forensic casework. Int J Legal Med 114:169–172
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Acknowledgments
We acknowledge Alexandra Lopes for collecting samples from central and south Portugal and providing them for this study. Luísa Pereira has a post-doctoral grant from Fundação para a Ciência e a Tecnologia (SFRH/BPD/7121/2001). IPATIMUP is supported by Programa Operacional Ciência, Tecnologia e Inovação (POCTI), Quadro Comunitário de Apoio III.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Pereira, L., Cunha, C. & Amorim, A. Predicting sampling saturation of mtDNA haplotypes: an application to an enlarged Portuguese database. Int J Legal Med 118, 132–136 (2004). https://doi.org/10.1007/s00414-003-0424-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-003-0424-1