Abstract
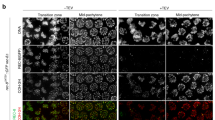
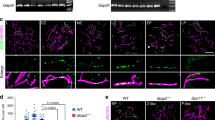
The meiotic recombination checkpoint blocks meiotic cell cycle progression in response to synapsis and/or recombination defects to prevent aberrant chromosome segregation. The evolutionarily conserved budding yeast Pch2TRIP13 AAA+ ATPase participates in this pathway by supporting phosphorylation of the Hop1HORMAD adaptor at T318. In the wild type, Pch2 localizes to synapsed chromosomes and to the unsynapsed rDNA region (nucleolus), excluding Hop1. In contrast, in synaptonemal complex (SC)–defective zip1Δ mutants, which undergo checkpoint activation, Pch2 is detected only on the nucleolus. Alterations in some epigenetic marks that lead to Pch2 dispersion from the nucleolus suppress zip1Δ-induced checkpoint arrest. These observations have led to the notion that Pch2 nucleolar localization could be important for the meiotic recombination checkpoint. Here we investigate how Pch2 chromosomal distribution impacts checkpoint function. We have generated and characterized several mutations that alter Pch2 localization pattern resulting in aberrant Hop1 distribution and compromised meiotic checkpoint response. Besides the AAA+ signature, we have identified a basic motif in the extended N-terminal domain critical for Pch2’s checkpoint function and localization. We have also examined the functional relevance of the described Orc1-Pch2 interaction. Both proteins colocalize in the rDNA, and Orc1 depletion during meiotic prophase prevents Pch2 targeting to the rDNA allowing unwanted Hop1 accumulation on this region. However, Pch2 association with SC components remains intact in the absence of Orc1. We finally show that checkpoint activation is not affected by the lack of Orc1 demonstrating that, in contrast to previous hypotheses, nucleolar localization of Pch2 is actually dispensable for the meiotic checkpoint.







Similar content being viewed by others
References
Acosta I, Ontoso D, San-Segundo PA (2011) The budding yeast polo-like kinase Cdc5 regulates the Ndt80 branch of the meiotic recombination checkpoint pathway. Mol Biol Cell 22:3478–3490
Bell SP, Kobayashi R, Stillman B (1993) Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844–1849
Bhalla N, Dernburg AF (2005) A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310:1683–1686
Börner GV, Barot A, Kleckner N (2008) Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc Natl Acad Sci U S A 105:3327–3332
Carballo JA, Johnson AL, Sedgwick SG, Cha RS (2008) Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132:758–770
Cavero S, Herruzo E, Ontoso D, San-Segundo PA (2016) Impact of histone H4K16 acetylation on the meiotic recombination checkpoint in Saccharomyces cerevisiae. Microb Cell 3:606–620
Chakraborty P, Pankajam AV, Lin G, Dutta A, Krishnaprasad GN, Tekkedil MM, Shinohara A, Steinmetz LM, Nishant KT (2017) Modulating crossover frequency and interference for obligate crossovers in Saccharomyces cerevisiae meiosis. G3 7:1511–1524
Chen C, Jomaa A, Ortega J, Alani EE (2014) Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc Natl Acad Sci U S A 111:E44–E53
Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF (2006) SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev 20:2067–2081
Deshong AJ, Ye AL, Lamelza P, Bhalla N (2014) A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet 10:e1004291
Dong H, Roeder GS (2000) Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol 148:417–426
Duncker BP, Chesnokov IN, McConkey BJ (2009) The origin recognition complex protein family. Genome Biol 10:214
Eichinger CS, Jentsch S (2010) Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc Natl Acad Sci U S A 107:11370–11375
Farmer S, Hong EJ, Leung WK, Argunhan B, Terentyev Y, Humphryes N, Toyoizumi H, Tsubouchi H (2012) Budding yeast Pch2, a widely conserved meiotic protein, is involved in the initiation of meiotic recombination. PLoS One 7:e39724
Fox CA, Loo S, Dillin A, Rine J (1995) The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev 9:911–924
Fox C, Zou J, Rappsilber J, Marston AL (2017) Cdc14 phosphatase directs centrosome re-duplication at the meiosis I to meiosis II transition in budding yeast. Wellcome Open Res 2:2
Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553
Gottlieb S, Esposito RE (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776
Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL (2010) Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev 24:1772–1786
Hanner AS, Rusche LN (2017) The yeast heterochromatin protein Sir3 experienced functional changes in the AAA+ domain after gene duplication and subfunctionalization. Genetics 207:517–528
Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6:519–529
Herruzo E, Ontoso D, Gonzalez-Arranz S, Cavero S, Lechuga A, San-Segundo PA (2016) The Pch2 AAA+ ATPase promotes phosphorylation of the Hop1 meiotic checkpoint adaptor in response to synaptonemal complex defects. Nucleic Acids Res 44:7722–7741
Ho HC, Burgess SM (2011) Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet 7:e1002351
Hong EJ, Roeder GS (2002) A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev 16:363–376
Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962
Joshi N, Barot A, Jamison C, Börner GV (2009) Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet 5:e1000557
Joshi N, Brown MS, Bishop DK, Börner GV (2015) Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol Cell 57:797–811
Joyce EF, McKim KS (2009) Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics 181:39–51
Joyce EF, McKim KS (2010) Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet 6:e1001059
Kniewel R, Murakami H, Liu Y, Ito M, Ohta K, Hollingsworth NM, Keeney S (2017) Histone H3 threonine 11 phosphorylation is catalyzed directly by the meiosis-specific kinase Mek1 and provides a molecular readout of Mek1 activity in vivo. Genetics 207:1313–1333
Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, Yang J, Armstrong SJ, Mechtler K, Roitinger E, Franklin FC (2015) Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet 11:e1005372
Li XC, Schimenti JC (2007) Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet 3:e130
Lydall D, Nikolsky Y, Bishop DK, Weinert T (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840–843
Ma HT, Poon RYC (2018) TRIP13 functions in the establishment of the spindle assembly checkpoint by replenishing O-MAD2. Cell Rep 22:1439–1450
Medhi D, Goldman AS, Lichten M (2016) Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. Elife 5:e19699
Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z (2013) Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25:2998–3009
Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M Jr (2010) Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev 24:2693–2704
Nelson CR, Hwang T, Chen PH, Bhalla N (2015) TRIP13PCH-2 promotes Mad2 localization to unattached kinetochores in the spindle checkpoint response. J Cell Biol 211:503–516
Nishimura K, Kanemaki MT (2014) Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID). Curr Protoc Cell Biol 64:20.9.1–20.916
Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS (2016) Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol Cell 64:405–415
Ontoso D, Acosta I, van Leeuwen F, Freire R, San-Segundo PA (2013) Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. PLoS Genet 9:e1003262
Penedos A, Johnson AL, Strong E, Goldman AS, Carballo JA, Cha RS (2015) Essential and checkpoint functions of budding yeast ATM and ATR during meiotic prophase are facilitated by differential phosphorylation of a meiotic adaptor protein, Hop1. PLoS One 10:e0134297
Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I (2005) The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci 118:4261–4269
Prugar E, Burnett C, Chen X, Hollingsworth NM (2017) Coordination of double strand break repair and meiotic progression in yeast by a Mek1-Ndt80 negative feedback loop. Genetics 206:497–512
Qiu ZR, Shuman S, Schwer B (2011) An essential role for trimethylguanosine RNA caps in Saccharomyces cerevisiae meiosis and their requirement for splicing of SAE3 and PCH2 meiotic pre-mRNAs. Nucleic Acids Res 39:5633–5646
Refolio E, Cavero S, Marcon E, Freire R, San-Segundo PA (2011) The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J Cell Sci 124:2488–2500
Rockmill B (2009) Chromosome spreading and immunofluorescence methods in Saccharomyes cerevisiae. Methods Mol Biol 558:3–13
Rockmill B, Roeder GS (1990) Meiosis in asynaptic yeast. Genetics 126:563–574
Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S (2010) Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet 6:1001062
San-Segundo PA, Roeder GS (1999) Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97:313–324
San-Segundo PA, Roeder GS (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell 11:3601–3615
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682
Smith AV, Roeder GS (1997) The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136:957–967
Stegmeier F, Amon A (2004) Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38:203–232
Stuckey S, Mukherjee K, Storici F (2011) In vivo site-specific mutagenesis and gene collage using the delitto perfetto system in yeast Saccharomyces cerevisiae. Methods Mol Biol 745:173–191
Subramanian VV, Hochwagen A (2014) The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6:a016675
Subramanian VV, MacQueen AJ, Vader G, Shinohara M, Sanchez A, Borde V, Shinohara A, Hochwagen A (2016) Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol 14:e1002369
Sym M, Engebrecht JA, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72:365–378
Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7:1255–1266
Vader G (2015) Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma 124:333–339
Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A (2011) Protection of repetitive DNA borders from self-induced meiotic instability. Nature 477:115–119
Villoria MT, Ramos F, Duenas E, Faull P, Cutillas PR, Clemente-Blanco A (2017) Stabilization of the metaphase spindle by Cdc14 is required for recombinational DNA repair. EMBO J 36:79–101
Voelkel-Meiman K, Moustafa SS, Lefrancois P, Villeneuve AM, MacQueen AJ (2012) Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genet 8:e1002993
Wendler P, Ciniawsky S, Kock M, Kube S (2012) Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823:2–14
West AMV, Komives EA, Corbett KD (2018) Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res 46:279–292
Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5:e1000702
Wu HY, Burgess SM (2006) Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol 16:2473–2479
Xie B, Horecka J, Chu A, Davis RW, Becker E, Primig M (2016) Ndt80 activates the meiotic ORC1 transcript isoform and SMA2 via a bi-directional middle sporulation element in Saccharomyces cerevisiae. RNA Biol 13:772–782
Ye Q, Rosenberg SC, Moeller A, Speir JA, Su TY, Corbett KD (2015) TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife 4:e07367
Zanders S, Alani E (2009) The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet 5:e1000571
Zanders S, Sonntag Brown M, Chen C, Alani E (2011) Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics 188:511–521
Acknowledgements
We are grateful to David Ontoso, Andrés Clemente, and Shirleen Roeder for reagents. We also thank Isabel Acosta and Sara González-Arranz for the technical assistance, Carlos Vázquez for the advice on microscopy analysis, and José Pérez-Martín and Andrés Clemente for the helpful discussions and ideas.
Funding
This work was supported by grants from the Ministry of Economy and Competitiveness (MINECO) of Spain to JAC and PSS (grants BFU2015-64361-P and BFU2015-65417-R, respectively). EH was supported by a predoctoral contract (FPU1502035) from the Ministry of Education of Spain. JAC is supported by a Ramón y Cajal contract (RYC2013-13950). The IBFG is funded in part by an institutional grant from Junta de Castilla y León (CLU-2017-03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of a Special Issue on Recent advances in meiosis from DNA replication to chromosome segregation “edited by Valérie Borde and Francesca Cole, co-edited by Paula Cohen and Scott Keeney.”
Electronic supplementary material
ESM 1
(PDF 2571 kb)
Rights and permissions
About this article
Cite this article
Herruzo, E., Santos, B., Freire, R. et al. Characterization of Pch2 localization determinants reveals a nucleolar-independent role in the meiotic recombination checkpoint. Chromosoma 128, 297–316 (2019). https://doi.org/10.1007/s00412-019-00696-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00696-7