Abstract
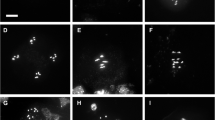
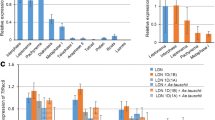
In contrast to yeast, plant interphase nuclei often display incomplete alignment (cohesion) along sister chromatid arms. Sister chromatid cohesion mediated by the multi-subunit cohesin complex is essential for correct chromosome segregation during nuclear divisions and for DNA recombination repair. The cohesin complex consists of the conserved proteins SMC1, SMC3, SCC3, and an α-kleisin subunit. Viable homozygous mutants could be selected for the Arabidopsis thaliana α-kleisins SYN1, SYN2, and SYN4, which can partially compensate each other. For the kleisin SYN3 and for the single-copy genes SMC1, SMC3, and SCC3, only heterozygous mutants were obtained that displayed between 77% and 97% of the wild-type transcript level. Compared to wild-type nuclei, sister chromatid alignment was significantly decreased along arms in 4C nuclei of the homozygous syn1 and syn4 and even of the heterozygous smc1, smc3, scc3, and syn3 mutants. Knocking out SYN1 and SYN4 additionally impaired sister centromere cohesion. Homozygous mutants of SWITCH1 (required for meiotic sister chromatid alignment) displayed sterility and decreased sister arm alignment. For the cohesin loading complex subunit SCC2, only heterozygous mutants affecting sister centromere alignment were obtained. Defects of the α-kleisin SYN4, which impair sister chromatid alignment in 4C differentiated nuclei, do apparently not disturb alignment during prometaphase nor cause aneuploidy in meristematic cells. The syn2, 3, 4 scc3 and swi1 mutants display a high frequency of anaphases with bridges (~10% to >20% compared to 2.6% in wild type). Our results suggest that (a) already a slight reduction of the average transcript level in heterozygous cohesin mutants may cause perturbation of cohesion, at least in some leaf cells at distinct loci; (b) the decreased sister chromatid alignment in cohesin mutants can obviously not fully be compensated by other cohesion mechanisms such as DNA concatenation; (c) some cohesin genes, in addition to cohesion, might have further essential functions (e.g., for genome stability, apparently by facilitating correct recombination repair of double-strand breaks).




Similar content being viewed by others
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11:417–430
Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL (2007) Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol 27:8522–8532
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542
Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat J-P (2008) Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J 27:111–121
Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I (2006) Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J 48:771–783
Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19:463–472
Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249–259
Cai X, Dong F, Edelmann RE, Makaroff CA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116:2999–3007
Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S (2007) Protein requirements for sister telomere association in human cells. EMBO J 26:4867–4878
Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mézard C, Grelon M (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118:4621–4632
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5:243–254
Cortés-Ledesma F, Aguilera A (2006) Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7:919–926
Cortés-Ledesma F, de Piccoli G, Haber JE, Aragón L, Aguilera A (2007) SMC proteins, new players in the maintenance of genomic stability. Cell Cycle 6:914–918
da Costa-Nunes JA, Bhatt AM, O'Shea S, West CE, Bray CM, Grossniklaus U, Dickinson HG (2006) Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J Exp Bot 57:971–983
Dej KJ, Ahn C, Orr-Weaver TL (2004) Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics 168:895–906
Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ (2008) Chromosome cohesion—rings, knots, orcs and fellowship. J Cell Sci 121:2107–2114
Dong F, Cai X, Makaroff CA (2001) Cloning and characterization of two Arabidopsis genes that belong to the RAD21/REC8 family of chromosome cohesin proteins. Gene 271:99–108
Dorsett D (2007) Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116:1–13
Fagan T (1996) QUICKBASIC program for exact and mid-p confidence interval for a binomial proportion. Comput Biol Med 26:263–267
Gause M, Schaaf CA, Dorsett D (2008) Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays 30:715–718
Glynn EF, Megee PC, Yu HG, Mistrot C, Ünal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2:1325–1339
Guacci V (2007) Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells 12:693–708
Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125:517–530
Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132:983–995
Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21:3144–3158
Heidinger-Pauli JM, Ünal E, Guacci V, Koshland D (2008) The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell 31:47–56
Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF (2004) The Drosophila meiotic kleisin C(2) M functions before the meiotic divisions. Chromosoma 113:177–187
Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7:311–322
Jiang L, Xia M, Strittmatter LI, Makaroff CA (2007) The Arabidopsis cohesin protein SYN3 localizes to the nucleolus and is essential for gametogenesis. Plant J 50:1020–1034
Kawabe A, Nasuda S (2005) Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol Genet Genomics 272:593–602
Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K (2002) Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem 277:45149–45153
Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151:1047–1056
Lam WS, Yang X, Makaroff CA (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci 118:3037–3048
Lam WW, Peterson EA, Yeung M, Lavoie BD (2006) Condensin is required for chromosome arm cohesion during mitosis. Genes Dev 20:2973–2984
Lee JY, Orr-Weaver TL (2001) The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol 17:753–777
Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578
Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23:787–799
Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29:405–415
Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116:321–329
Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19:1269–1287
Maguire MP (1990) Sister chromatid cohesiveness: vital function, obscure mechanism. Biochem Cell Biol 68:1231–1242
Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefèvre F, Pelletier G, Horlow C (2001) SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15:1859–1871
Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FC (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130:3309–3318
Merkle CJ, Karnitz LM, Henry-Sánchez JT, Chen J (2003) Cloning and characterization of hCTF18, hCTF8, and hDCC1—human homologs of an Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J Biol Chem 278:30051–30056
Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D (2008) Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117:89–102
Mito Y, Sugimoto A, Yamamoto M (2003) Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol Biol Cell 14:2399–2409
Miyazaki WY, Orr-Weaver TL (1994) Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28:167–187
Murray JM, Carr AM (2008) SMC5/6: a link between DNA repair and undirectional replication? Nat Rev Mol Cell Biol 9:177–182
Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F (2007) Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma 116:531–544
Ogiwara H, Ohuchi T, Ui A, Tada S, Enomoto T, Seki M (2007) Ctf18 is required for homologous recombination-mediated double-strand break repair. Nucleic Acids Res 35:4989–5000
Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24:105–129
Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschiager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15:1349–1360
Pasierbek P, Födermayr M, Jantsch V, Jantsch M, Schweizer D, Loidl J (2003) The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp Cell Res 289:245–255
Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269
Peric-Hupkes D, van Steensel B (2008) Linking cohesin to gene regulation. Cell 132:925–928
Potts PR, Porteus MH, Yu HT (2006) Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25:3377–3388
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Schubert V (2009) SMC proteins and their multiple functions in higher plants. Cytogenet Genome Res (in press)
Schubert I, Fransz PF, Fuchs J, de Jong JH (2001) Chromosome painting in plants. Methods Cell Sci 23:57–69
Schubert V, Klatte M, Pecinka A, Meister A, Jasencakova Z, Schubert I (2006) Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172:467–475
Schubert V, Kim YM, Berr A, Fuchs J, Meister A, Marschner S, Schubert I (2007) Random homologous pairing and incomplete sister chromatid alignment are common in angiosperm interphase nuclei. Mol Genet Genomics 278:167–176
Schubert V, Kim YM, Schubert I (2008) Arabidopsis sister chromatids often show complete alignment or separation along a 1.2-Mb euchromatic region but no cohesion “hot spots”. Chromosoma 117:261–266
Sebastian J, Ravi M, Andreuzza S, Panoli AP, Marimuthu MPA, Siddiqi I (2009) The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in Arabidopsis. Plant J . doi:10.1111/j.1365-313X.2009.03845.x
Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T (2006) Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol 4:1411–1425
Selig S, Okumura K, Ward DC, Cedar H (1992) Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J 11:1217–1225
Skibbens RV, Maradeo M, Eastman L (2007) Fork it over: the cohesion establishment factor Ctf7p and DNA replication. J Cell Sci 120:2471–2477
Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666
Ström L, Sjögren C (2007) Chromosome segregation and double-strand break repair—a complex connection. Curr Opin Cell Biol 19:344–349
Ström L, Lindroos HB, Shirahige K, Sjögren C (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16:1003–1015
Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C (2007) Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317:242–245
Sundin O, Varshavsky A (1980) Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21:103–114
Tanaka T, Cosma MP, Wirth K, Nasmyth K (1999) Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98:847–858
Tao J, Zhang L, Chong K, Wang T (2007) OsRAD21–3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J 51:919–930
Uhlmann F (2008) Molecular biology: cohesin branches out. Nature 451:777–778
Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D (2004) DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16:991–1002
Ünal E, Heidinger-Pauli JM, Koshland D (2007) DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317:245–248
Vas AC, Andrews CA, Kirkland Matesky K, Clarke DJ (2007) In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Mol Biol Cell 18:557–568
Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM (2003) Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol 13:208–218
Volpi EV, Sheer D, Uhlmann F (2001) Cohesion, but not too close. Curr Biol 11:R378
Wang F, Yoder J, Antoshechkin I, Han M (2003) Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol Cell Biol 23:7698–7707
Ward PB (2002) FISH probes and labelling techniques. In: Beatty B, Mai S, Squire J (eds) Fish. Oxford University, Oxford, pp 5–28
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM (2006) Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16:863–874
Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC (2004) The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol 2:1340–1353
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801
Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol 18:81–90
Zhang LR, Tao JY, Wang T (2004) Molecular characterization of OsRAD21–1, a rice homologue of yeast RAD21 essential for mitotic chromosome cohesion. J Exp Bot 55:1149–1152
Zhang L, Tao J, Wang S, Chong K, Wang T (2006) The rice OsRad21–4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60:533–554
Acknowledgments
We thank Martina Kühne, Joachim Bruder, and Rita Schubert for technical assistance and Andreas Houben for critical reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Uhlmann
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Table S1
PCR primers used to identify the T-DNA insertion alleles. (DOC 14 kb)
Table S2
Sequences of the left border junctions of the T-DNA insertion lines. (DOC 13 kb)
Table S3
RT and real-time PCR primers used to amplify transcripts. (DOC 14 kb)
Rights and permissions
About this article
Cite this article
Schubert, V., Weißleder, A., Ali, H. et al. Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana . Chromosoma 118, 591–605 (2009). https://doi.org/10.1007/s00412-009-0220-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0220-x