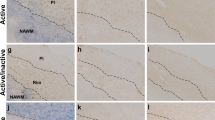
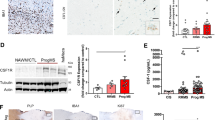
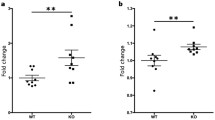
Abstract
Myeloid-derived cells play important modulatory and effector roles in multiple sclerosis (MS) and its animal model, experimental autoimmune encephalomyelitis (EAE). Myeloid-derived suppressor cells (MDSCs) are immature myeloid cells, composed of monocytic (MO) and polymorphonuclear (PMN) fractions, which can suppress T cell activities in EAE. Their role in MS remains poorly characterized. We found decreased numbers of circulating MDSCs, driven by lower frequencies of the MO-MDSCs, and higher MDSC expression of microRNA miR-223 in MS versus healthy subjects. To gain mechanistic insights, we interrogated the EAE model. MiR-223 knock out (miR-223−/−) mice developed less severe EAE with increased MDSC numbers in the spleen and spinal cord compared to littermate controls. MiR-223−/− MO-MDSCs suppressed T cell proliferation and cytokine production in vitro and EAE in vivo more than wild-type MO-MDSCs. They also displayed an increased expression of critical mediators of MDSC suppressive function, Arginase-1(Arg1), and the signal transducer and activator of transcription 3 (Stat3), which herein, we demonstrate being an miR-223 target gene. Consistently, MDSCs from MS patients displayed decreased STAT3 and ARG1 expression compared with healthy controls, suggesting that circulating MDSCs in MS are not only reduced in numbers but also less suppressive. These results support a critical role for miR-223 in modulating MDSC biology in EAE and in MS and suggest potential novel therapeutic applications.








Similar content being viewed by others
Abbreviations
- MS:
-
Multiple sclerosis
- miRNAs:
-
MicroRNAs
- MDSCs:
-
Myeloid-derived suppressor cells
- EAE:
-
Experimental autoimmune encephalomyelitis
- STAT3:
-
Signal transducer and activator of transcription 3
- ARG1:
-
Arginase-1
References
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. doi:10.1038/nature02871
Bader AG, Brown D, Winkler M (2010) The promise of microRNA replacement therapy. Cancer Res 70:7027–7030. doi:10.1158/0008-5472.CAN-10-2010
Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK (2003) Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med 197:1073–1081. doi:10.1084/jem.20021603
Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S et al (2016) Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 7:12150. doi:10.1038/ncomms12150
Condamine T, Ramachandran I, Youn JI, Gabrilovich DI (2015) Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med 66:97–110. doi:10.1146/annurev-med-051013-052304
De Santis G, Ferracin M, Biondani A, Caniatti L, Rosaria Tola M, Castellazzi M, Zagatti B, Battistini L, Borsellino G, Fainardi E et al (2010) Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol 226:165–171. doi:10.1016/j.jneuroim.2010.06.009
Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F et al (2007) Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12:457–466. doi:10.1016/j.ccr.2007.09.020
Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123:819–831. doi:10.1016/j.cell.2005.09.023
Fenoglio C, Ridolfi E, Cantoni C, De Riz M, Bonsi R, Serpente M, Villa C, Pietroboni AM, Naismith RT, Alvarez E et al (2013) Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult Scler 19:1938–1942. doi:10.1177/1352458513485654
Fletcher JM, Lalor SJ, Sweeney CM, Tubridy N, Mills KH (2010) T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin Exp Immunol 162:1–11. doi:10.1111/j.1365-2249.2010.04143.x
Gabrilovich DI, Ostrand-Rosenberg S, Bronte V (2012) Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 12:253–268. doi:10.1038/nri3175
Gilicze AB, Wiener Z, Toth S, Buzas E, Pallinger E, Falcone FH, Falus A (2014) Myeloid-derived microRNAs, miR-223, miR27a, and miR-652, are dominant players in myeloid regulation. Biomed Res Int 2014:870267. doi:10.1155/2014/870267
Gimenez MA, Sim J, Archambault AS, Klein RS, Russell JH (2006) A tumor necrosis factor receptor 1-dependent conversation between central nervous system-specific T cells and the central nervous system is required for inflammatory infiltration of the spinal cord. Am J Pathol 168:1200–1209. doi:10.2353/ajpath.2006.050332
Guerau-de-Arellano M, Alder H, Ozer HG, Lovett-Racke A, Racke MK (2012) miRNA profiling for biomarker discovery in multiple sclerosis: from microarray to deep sequencing. J Neuroimmunol 248:32–39. doi:10.1016/j.jneuroim.2011.10.006
Guerau-de-Arellano M, Liu Y, Meisen WH, Pitt D, Racke MK, Lovett-Racke AE (2015) Analysis of miRNA in normal appearing white matter to identify altered CNS pathways in multiple sclerosis. J Autoimmune Disord 1:1–8
Hoechst B, Gamrekelashvili J, Manns MP, Greten TF, Korangy F (2011) Plasticity of human Th17 cells and iTregs is orchestrated by different subsets of myeloid cells. Blood 117:6532–6541. doi:10.1182/blood-2010-11-317321
Ifergan I, Chen S, Zhang B, Miller SD (2016) Cutting edge: microRNA-223 regulates myeloid dendritic cell-driven Th17 responses in experimental autoimmune encephalomyelitis. J Immunol 196:1455–1459. doi:10.4049/jimmunol.1501965
Ioannou M, Alissafi T, Lazaridis I, Deraos G, Matsoukas J, Gravanis A, Mastorodemos V, Plaitakis A, Sharpe A, Boumpas D et al (2012) Crucial role of granulocytic myeloid-derived suppressor cells in the regulation of central nervous system autoimmune disease. J Immunol 188:1136–1146. doi:10.4049/jimmunol.1101816
Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK (2009) Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol 183:7169–7177. doi:10.4049/jimmunol.0901906
Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451:1125–1129. doi:10.1038/nature06607
Junker A, Hohlfeld R, Meinl E (2011) The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol 7:56–59. doi:10.1038/nrneurol.2010.179
Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E (2009) MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain J Neurol 132:3342–3352. doi:10.1093/brain/awp300
Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, Lenhof HP, Ruprecht K, Meese E (2009) Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 4:e7440. doi:10.1371/journal.pone.0007440
Kiernan JA (1984) Chromoxane cyanine R. II. Staining of animal tissues by the dye and its iron complexes. J Microsc 134:25–39
King IL, Dickendesher TL, Segal BM (2009) Circulating Ly-6C+ myeloid precursors migrate to the CNS and play a pathogenic role during autoimmune demyelinating disease. Blood 113:3190–3197. doi:10.1182/blood-2008-07-168575
Kotsakis A, Harasymczuk M, Schilling B, Georgoulias V, Argiris A, Whiteside TL (2012) Myeloid-derived suppressor cell measurements in fresh and cryopreserved blood samples. J Immunol Methods 381:14–22. doi:10.1016/j.jim.2012.04.004
Kumar V, Patel S, Tcyganov E, Gabrilovich DI (2016) The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol 37:208–220. doi:10.1016/j.it.2016.01.004
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. doi:10.1016/j.cell.2004.12.035
Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG (2010) MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11:799–805. doi:10.1038/ni.1918
Liu Q, Zhang M, Jiang X, Zhang Z, Dai L, Min S, Wu X, He Q, Liu J, Zhang Y et al (2011) miR-223 suppresses differentiation of tumor-induced CD11b(+) Gr1(+) myeloid-derived suppressor cells from bone marrow cells. Int J Cancer 129:2662–2673. doi:10.1002/ijc.25921
Moline-Velazquez V, Cuervo H, Vila-Del Sol V, Ortega MC, Clemente D, de Castro F (2011) Myeloid-derived suppressor cells limit the inflammation by promoting T lymphocyte apoptosis in the spinal cord of a murine model of multiple sclerosis. Brain Pathol 21:678–691. doi:10.1111/j.1750-3639.2011.00495.x
Moline-Velazquez V, Vila-Del Sol V, de Castro F, Clemente D (2016) Myeloid cell distribution and activity in multiple sclerosis. Histol Histopathol 31:357–370. doi:10.14670/HH-11-699
Nagaraj S, Youn JI, Gabrilovich DI (2013) Reciprocal relationship between myeloid-derived suppressor cells and T cells. J Immunol 191:17–23. doi:10.4049/jimmunol.1300654
Novitskiy SV, Pickup MW, Gorska AE, Owens P, Chytil A, Aakre M, Wu H, Shyr Y, Moses HL (2011) TGF-beta receptor II loss promotes mammary carcinoma progression by Th17 dependent mechanisms. Cancer Discov 1:430–441. doi:10.1158/2159-8290.CD-11-0100
O’Connell RM, Zhao JL, Rao DS (2011) MicroRNA function in myeloid biology. Blood 118:2960–2969. doi:10.1182/blood-2011-03-291971
Parekh VV, Wu L, Olivares-Villagomez D, Wilson KT, Van Kaer L (2013) Activated invariant NKT cells control central nervous system autoimmunity in a mechanism that involves myeloid-derived suppressor cells. J Immunol 190:1948–1960. doi:10.4049/jimmunol.1201718
Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, Bronte V (2010) Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol 22:238–244. doi:10.1016/j.coi.2010.01.021
Piccio L, Cantoni C, Henderson JG, Hawiger D, Ramsbottom M, Mikesell R, Ryu J, Hsieh CS, Cremasco V, Haynes W et al (2013) Lack of adiponectin leads to increased lymphocyte activation and increased disease severity in a mouse model of multiple sclerosis. Eur J Immunol 43:2089–2100. doi:10.1002/eji.201242836
Piccio L, Stark JL, Cross AH (2008) Chronic calorie restriction attenuates experimental autoimmune encephalomyelitis. J Leukoc Biol 84:940–948. doi:10.1189/jlb.0208133
Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302. doi:10.1002/ana.22366
Procaccini C, De Rosa V, Pucino V, Formisano L, Matarese G (2015) Animal models of multiple sclerosis. Eur J Pharmacol 759:182–191. doi:10.1016/j.ejphar.2015.03.042
Ridolfi E, Fenoglio C, Cantoni C, Calvi A, De Riz M, Pietroboni A, Villa C, Serpente M, Bonsi R, Vercellino M et al (2013) Expression and genetic analysis of microRNAs involved in multiple sclerosis. Int J Mol Sci 14:4375–4384. doi:10.3390/ijms14034375
Rumble JM, Huber AK, Krishnamoorthy G, Srinivasan A, Giles DA, Zhang X, Wang L, Segal BM (2015) Neutrophil-related factors as biomarkers in EAE and MS. J Exp Med 212:23–35. doi:10.1084/jem.20141015
Sullivan RP, Leong JW, Schneider SE, Ireland AR, Berrien-Elliott MM, Singh A, Schappe T, Jewell BA, Sexl V, Fehniger TA (2015) MicroRNA-15/16 antagonizes Myb to control NK cell maturation. J Immunol 195:2806–2817. doi:10.4049/jimmunol.1500949
Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H et al (2013) STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Investig 123:1580–1589. doi:10.1172/JCI60083
Yi H, Guo C, Yu X, Zuo D, Wang XY (2012) Mouse CD11b+ Gr-1+ myeloid cells can promote Th17 cell differentiation and experimental autoimmune encephalomyelitis. J Immunol 189:4295–4304. doi:10.4049/jimmunol.1200086
Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY et al (2008) TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453:236–240. doi:10.1038/nature06878
Zhu B, Bando Y, Xiao S, Yang K, Anderson AC, Kuchroo VK, Khoury SJ (2007) CD11b+ Ly-6C(hi) suppressive monocytes in experimental autoimmune encephalomyelitis. J Immunol 179:5228–5237
Zhu B, Kennedy JK, Wang Y, Sandoval-Garcia C, Cao L, Xiao S, Wu C, Elyaman W, Khoury SJ (2011) Plasticity of Ly-6C(hi) myeloid cells in T cell regulation. J Immunol 187:2418–2432. doi:10.4049/jimmunol.1100403
Acknowledgments
We thank Anne H. Cross, MD for careful reading of the manuscript; Julia Sim and Angela Archambault, PhD for technical assistance and advices; Erin Longbrake, MD PhD for helping with patient enrollment in the study; all MS patients and healthy controls that donated blood for this project as well as the study coordinators that drew the blood: Samantha Lancia, Susan Fox, and Bridgette Clay. LP is a Harry Weaver Neuroscience Scholar of the National Multiple Sclerosis Society (NMSS, JF 2144A2/1) and supported by Fondazione Italiana Sclerosi Multipla (FISM; 2014/R/15). GFW was supported by R01NS083678. LP and GFW were funded by the Dana Foundation “Program in the Neuroimmunology and Brain Infections and Cancer”. CC was supported during the course of this study by a FISM fellowship (2012/B/1) and subsequently by a NMSS fellowship (FG 2010-A1/2). TAF was supported by R01AI102924. Patients were seen for this study in the Neuroclinical Research Unit (NCRU) supported by the National Institute of Health (CO6 RR020092) and Washington University Insitute of Clinical and Translational Sciences-Brain Behavioral and Performance Unit (TR000448).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2016_1621_MOESM1_ESM.tif
Supplementary Fig. 1. Percentages of MDSCs and MO-MDSCs in the peripheral blood of healthy control subjects (n = 16), untreated and GA-treated RRMS patients (n = 10 and n = 24). a, b Percentages of MDSCs and MO-MDSCs were significantly lower in untreated RRMS patients compared to healthy controls. GA-treated patient showed a modest increase in MDSC and MO-MDSC percentages, but this was not statistically significant. Percentages were calculated relative to a gate which included whole leukocytes. Error bars represent the mean ± SD. P values were calculated by Kruskal–Wallis H test and post hoc analysis (TIFF 9217 kb)
401_2016_1621_MOESM2_ESM.tif
Supplementary Fig. 2. Mir-223 expression in the CNS and lymphoid tissues in C57BL/6 mice during EAE. MiR-223 expression in spleen, brain, lymph nodes, and bone marrow taken from C57BL/6 naïve mice and on days 5, 10 15, and 30 post-EAE immunization (n = 4 mice/time point) analyzed by quantitative RT-PCR. U6 snRNA was used as endogenous controls (TIFF 44468 kb)
401_2016_1621_MOESM3_ESM.tif
Supplementary Fig. 3. Size and cellularity of WT and miR-223−/− lymph node draining the immunization. a At day 6, post-immunization WT and miR-223−/− lymph node draining the site of immunization were isolated and measured in size. b Scatter plot of the percentage of myeloid cells (defined as CD11b+Gr1+ cells) from WT and miR-223−/− lymph nodes. Error bars represent mean ± SEM. A representative experiment from a total of three performed is showed. P values were calculated by Mann–Whitney U test (TIFF 18680 kb)
401_2016_1621_MOESM4_ESM.tif
Supplementary Fig. 4. Absolute number of MO and PMN-MDSCs in naive WT and miR-223−/− mice. Cells were isolated from bone marrow, spleen, blood, and lymph nodes from naive WT and miR-223−/− mice and analyzed by flow cytometry. Gates were set on Zombie−CD45+CD11b+ cells; MO-MDSCs were defined as CD11b+Ly6Chigh cells, PMN-MDSCs as CD11b+Gr1+. Absolute numbers of MO and PMN-MDSCs were calculated from the percentages based on the total number of cells (bone marrow, spleen, and lymph nodes) or on the total number of cells per 100 μl of blood. Error bars represent the mean ± SEM. A representative experiment from a total of two is showed. P values were calculated by Mann–Whitney U test (TIFF 24193 kb)
401_2016_1621_MOESM5_ESM.tif
Supplementary Fig. 5. Characterization of surface markers expression and cytokine profile of MDSCs from immunized WT and miR-223−/− mice. a, b MO and PMN-MDSCs in the spleen and CNS-infiltrating cells from MOG35–55 immunized mice (15 DPI) were examined for the expression of MHC II, CD40, CD86, and PD-L1 by flow cytometry. Expression of individual markers is shown based on gating for MO- (CD11b+Ly6G−Ly6Chigh) and PMN-MDSCs (CD11b+Ly6G+Ly6Clow). Histograms for the different surface markers on MO and PMN-MDSCs in the spleen (a) and in the CNS (b) are referring to representative mice from each group. Red and blue lines indicate mean fluorescence intensity (MFI) in WT and miR-223−/−, respectively. Scatter plots are compiling the data from one experiment (four mice/group) shown as percentages of MO and PMN-MDSCs expressing the specific marker. c Analysis of cytokine (TNF-α, IL-10, and GM-CSF) production in splenic MO and PMN-MDSCs from WT and miR-223−/− mice at 15 DPI. Numbers indicate the percentage of cytokine-producing MO and PMN-MDSCs. Error bars are mean ± SEM. P values were calculated by Mann–Whitney U test (TIFF 153485 kb)
Rights and permissions
About this article
Cite this article
Cantoni, C., Cignarella, F., Ghezzi, L. et al. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol 133, 61–77 (2017). https://doi.org/10.1007/s00401-016-1621-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-016-1621-6