Abstract
Cardiac fibroblasts are the major producers of extracellular matrix (ECM) to form infarct scar. We hypothesized that fibroblasts undergo a spectrum of phenotype states over the course of myocardial infarction (MI) from early onset to scar formation. Fibroblasts were isolated from the infarct region of C57BL/6J male mice (3–6 months old, n = 60) at days 0 (no MI control) and 1, 3, or 7 after MI. Whole transcriptome analysis was performed by RNA-sequencing. Of the genes sequenced, 3371 were differentially expressed after MI. Enrichment analysis revealed that MI day 1 fibroblasts displayed pro-inflammatory, leukocyte-recruiting, pro-survival, and anti-migratory phenotype through Tnfrsf9 and CD137 signaling. MI day 3 fibroblasts had a proliferative, pro-fibrotic, and pro-angiogenic profile with elevated Il4ra signaling. MI day 7 fibroblasts showed an anti-angiogenic homeostatic-like myofibroblast profile and with a step-wise increase in Acta2 expression. MI day 7 fibroblasts relied on Pik3r3 signaling to mediate Tgfb1 effects and Fgfr2 to regulate PI3K signaling. In vitro, the day 3 MI fibroblast secretome stimulated angiogenesis, while day 7 MI fibroblast secretome repressed angiogenesis through Thbs1 signaling. Our results reveal novel mechanisms for fibroblasts in expressing pro-inflammatory molecules and regulating angiogenesis following MI.
Similar content being viewed by others
Introduction
Myocardial infarction (MI) occurs when coronary blood flow to the left ventricle (LV) is blocked for a sufficient duration to result in tissue necrosis. The cardiac wound healing response involves robust inflammation, extracellular matrix (ECM) remodeling, revascularization, and scar formation [23, 41, 53]. While the cardiac fibroblast serves a baseline homeostasis role or transforms to an activated myofibroblast in response to injury, intermediary phenotypes exist, and fibroblasts display a range of phenotypes in response to individual in vitro stimuli [1, 61]. For example, cardiac fibroblasts stimulated with lipopolysaccharide show increased pro-inflammatory cytokine production and decreased collagen synthesis [37, 59]. Fibroblasts stimulated with transforming growth factor (TGF)β1 increase collagen synthesis and differentiate to a myofibroblast phenotype [62, 65].
Angiogenesis is the formation of new microvessels that sprout from pre-existing endothelial cells to re-establish blood flow to the infarcted myocardium, making it a promising therapeutic strategy for MI [7]. While endothelial cells can self-activate to promote proliferation after injury, other cell types such as macrophages mediate endothelial cell activation and the formation of new capillaries [7]. Anti-inflammatory macrophages secrete factors that promote proliferation of both fibroblasts (TGFβ1) and endothelial cells (vascular endothelial growth factor, VEGF) to form granulation tissue [3]. The role of cardiac fibroblasts in regulating angiogenesis in the heart has not been thoroughly investigated.
The goals of this study were to combine transcriptomics and cell physiology to provide a map of fibroblast phenotypes in response to MI and to determine the mechanisms of the infarct fibroblast in mediating inflammation and angiogenesis. We analyzed transcriptomic changes at MI days 1, 3, and 7 which represented the early inflammatory, proliferative, and maturation phases of cardiac wound healing. We hypothesized that fibroblasts undergo a spectrum of phenotype states over the course of MI from early onset to scar formation. This cell phenotype range begins with resident cells maintaining homeostasis and ends with a new type of resident cell that maintains the MI reset in homeostasis. To our knowledge, this is the first study to detail the full transcriptome changes that occur in cardiac fibroblasts to mediate MI inflammation and angiogenesis.
Materials and methods (detailed methods in supplemental materials)
Animal use
All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of Mississippi Medical Center. A total of 69 C57BL/6 J adult (3–6 months old) male mice were included, 60 for transcriptome analysis, and 9 for multiplex immunofluorescence. Groups were randomly assigned prior to the surgeries by one investigator (KYDP) and another investigator (YM) performed the majority of surgeries. To reduce animal use as recommended by the National Centre for the Replacement, Refinement, and Reduction of Animals in Research [46], the fibroblasts used in this study were obtained from a subset of the same mice used for macrophage isolation [36].
Coronary artery ligation
To produce permanent MI, mice underwent coronary artery ligation surgery as described previously and according to the Guidelines for Experimental Models of Ischemia and Infarction [10, 21, 24, 31, 36]. Mice were anesthetized with 2% isoflurane, intubated, and ventilated. The left coronary artery was ligated with 8-0 suture, and MI was confirmed by LV blanching and ST-segment elevation on the EKG. Buprenorphine (0.5 mg/kg body weight) was administered to the mice immediately before surgery. Mice not receiving surgery were used as no MI controls (day 0), as sham-operated mice do not differ in physiology or inflammatory gene expression from non-surgical controls [18].
Echocardiography and necropsy
LV physiology was determined by transthoracic echocardiography (Vevo 2100, VisualSonics; Toronto, CA) as described before and according to the Guidelines for Measuring Cardiac Physiology in Mice [10, 21, 27, 31, 36]. Mice were anesthetized under 1–2% isoflurane, and both long- and short-axis images were obtained. Measurements were taken on the terminal day and were averaged from three cardiac cycles for each mouse. Following imaging, the hearts were removed and the left ventricle (LV) divided into remote and infarct (which included border zone) regions. Each region was separately weighed for infarct area estimation.
Isolation of LV infarct fibroblasts
LV fibroblasts were isolated from the infarct by immunomagnetic separation as described previously [10, 21, 36]. Excised LV tissue was rinsed and immediately minced and digested by collagenase II (Worthington; Lakewood, NJ) and DNase solution in Hanks-buffered saline solution. After digestion, a single-cell suspension was generated and filtered through a 30 µm pre-separation column. Cell suspensions were incubated at 4 °C for 10 min with an anti-Ly6G-biotin antibody followed by anti-biotin microbeads contained in the anti-Ly-6G microbead kit (#130-092-332; Miltenyi Biotech; Bergisch Gladbach, Germany) to remove neutrophils. Cells were incubated with anti-CD11b-microbeads (Miltenyi #130-049-601) for 15 min to remove macrophages. Cells conjugated to the antibody microbeads were separated by magnetic columns (Miltenyi #130-042-201), and the effluent (Ly6G- and Cd11b- cells) was plated in T25 flasks in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1× antibiotic–antimycotic solution (Gibco #15240-062). Fibroblasts at passage 2 were used for RNA-Sequencing (RNA-seq), proliferation assays, and secretome collection; fibroblasts at passage 3 were used for the migration assay. For cell and secretome collection, 4 × 104 cells were seeded into 6-well plates and grown to 80% confluence in DMEM supplemented with 10% fetal bovine serum (FBS). Cells were incubated in DMEM with 0.1% FBS for 16 h, after which the media was replaced with fresh media and collected after 24 h. The cell fraction was collected for RNA-seq, and the secretome was collected for mass spectrometry and immunoblotting examination and endothelial cell stimulation experiments.
RNA-seq
Fibroblasts were pooled to obtain three biological replicates for each day. Sample sizes were: day 0-n = 10 mice/pool; day 1-n = 3–6 mice/pool, day 3-n = 3 mice/pool, and day 7-n = 2–3 mice/pool. Whole transcriptome analysis was performed as described previously [21, 31, 36]. RNA was extracted using the Pure Link RNA Mini Kit (Ambion; Foster City, CA, USA) according to the manufacturer’s instructions and assessed for quality control parameters of minimum concentration and size range. The cDNA libraries were developed using the TruSeq Total Stranded RNA with RiboZero Kit (Ambion), Set-A, quantified with the Qubit System (Invitrogen; Carlsbad, CA, USA), and assessed for quality and size with the Experion DNA 1 K Chip (Bio-Rad; Hercules, CA). All samples passed quality control standards of minimum concentration and RNA Quality Indicator > 9, with discrete 18S and 28S bands. The library of n = 12 pooled samples was sequenced using the NextSeq 500 High Output Kit (300 cycles, paired end 100 bp) on the Illumina NextSeq 500 platform (Illumina; San Diego, CA, USA). Sequenced reads and Fastq sequence files were used to align reads to the reference genome USCS-GRCm38/mm10 in the RNA-Seq Alignment Application with STAR aligner using Illumina Cloud Computing Platform. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values of reference genes and transcripts were generated using Cufflinks 2.
Bioinformatic analyses
Analyses tools available in the online resource Metaboanalyst 3.0 (http://www.metaboanalyst.ca/) and GraphPad Prism were used for graphical and statistical analyses. FPKM values were uploaded into Metaboanalyst, and one-way ANOVA with Tukey’s post hoc test was performed to determine differentially expressed genes (defined as p < 0.05). Important feature analysis was used to identify the top five ranked genes at each MI day. Partial least-squares determinant analysis (PLS-DA) and heat mapping were used to evaluate changes in signaling genes. To assess differential expression at individual MI days, volcano plot analysis was performed comparing the MI day to day 0, with a cut-off fold-change threshold of > 1.2 and p < 0.05. A gene correlation network was constructed using the software Graphia Pro (Kajeka; Edinburgh, UK) and a Pearson correlation threshold of r > 0.95. The resultant graph was subjected to Markov clustering analysis to identify co-expression modules. Enrichment analysis for differentially expressed genes from each cluster was performed using Enrichr (http://amp.pharm.mssm.edu/Enrichr/) for both Panther Pathways and Gene Ontology (GO) Biological Processes. For MI signaling profiles, we performed partial least-squares discriminant analysis (PLS-DA) and heat mapping for receptors and intracellular signaling genes.
RT-PCR validation
Quantitative RT-PCR was used as a secondary validation of gene expression in the same samples used for RNA-Seq. Extracted RNA from cardiac fibroblasts was used to synthesize cDNA with the High Capacity RNA-to-cDNA kit (Applied Biosystems 4387406), and RT-PCR was performed using the Taqman Gene Expression Assay (Applied Biosystems). Acta2, Col3a1, and Postn were chosen as known fibroblast markers, while Cx3cl1 and Mmp14 were chosen to represent genes differentially expressed at different time points. Gene expression was calculated as 2−ΔCt to the housekeeping gene Hprt1 [26]. RT-PCR experiments were performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) with the exception that Hprt1 was used as the housekeeping gene [19].
Fibroblast cell physiology
Fibroblast proliferation was assessed by bromodeoxyuridine (BrdU) enzyme-linked immunosorbent assay (Roche Diagnostics #11647229001; Mannheim, Germany) as described previously using the same pooled samples for RNA-Seq at passage 2 [10, 21]. Cells were seeded into 96-well plates and 24 h later serum starved for 16–18 h. The plate was then incubated for 24 h in DMEM and antibiotics with either 0.1% or 10% FBS. Cells were incubated with BrdU (10 µM), fixed, and incubated with an anti-BrdU antibody. Absorbance at 370 nm and 492 nm was determined using a microplate reader, with measurements taken at 30 min and subtracting the limit value of 492 nm from 370 nm. Fibroblast migration was determined by electrical-cell impedance sensing (ECIS; Applied Biophysics) cells as described previously [10, 21]. Cells (4 × 104) were plated in 96-well plates with gold-film surface electrodes and cultured for 48 h. Cell monolayers were wounded, and rate of migration was assessed as change in impedance over time.
Multiplex immunofluorescence
LV mid-sections were fixed in 10% zinc-buffered formalin, paraffin-embedded, and sectioned at 5 µm. Slides were deparaffinized and rehydrated. Immunohistochemical staining was performed using the Opal 7-color Automation Kit (Perkin Elmer #NEL80100KT; Boston, MA, USA). Sections were stained with the following primary antibodies conjugated to Opal fluorophores: PDGFRα (1:100, R&D Systems; #AF1062; Minneapolis, MN) conjugated to Opal 520, and CX3CL1 (1:500; R&D #MAB571), CCL5/RANTES (R&D #AF478), VEGF (Abcam #ab51745), or thrombospondin 1 (Thbs1; 1:100; R&D Systems #AF3074) conjugated to Opal 650. Nuclei were counterstained with DAPI. Opal 520 and 650 fluorophores were visualized with FITC and Cy5 channels, respectively. Images were obtained using the Mantra System (Perkin Elmer) and quantified using inForm software (Perkin Elmer).
Mass spectrometry
Fibroblast secretomes (500 µl) from day 0 or MI day 7 were cleaned using Sep-Pak Vac C18 cartridge (Waters, Milford, MA), and then reduced, alkylated, and trypsin-digested into peptides. The peptides were cleaned again using another Sep-Pak Vac C18 cartridge (Waters, Milford, MA) and analyzed label-free by liquid chromatography–tandem mass spectrometry using a Q Exactive (ThermoFisher, Waltham, MA, USA). A 15 cm × 75 μm C18 column (5 μm particles with 100 Å pore size) was used and the nano-UPLC ran at 300 nL/min with a 150-min linear acetonitrile gradient (from 5 to 35% B over 150 min; A = 0.2% formic acid in water; B = 0.2% formic acid in 90% acetonitrile). Tandem mass spectrometry (MS/MS) was set with an exclusion of 25 s, and the samples were run with high-energy collisional dissociation fragmentation at normalized collision energy of 30 and an isolation width of 2 m/z. The resolution setting was 70,000 for target values of the MS at 1e6 ions and in MS2 at resolution setting of 17,500 for 1x105 ions. The identified peptides were quantified using spectral counting and total spectral counts of each sample were used for normalization, and a sample report was generated in Scaffold version 4.8.7 (Proteome Software, Portland, OR). Data (quantitative values of normalized total spectra) were analyzed by t test. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011778 [64].
Immunoblotting
Immunoblotting was performed according to the Guidelines for authors and reviewers on antibody use in physiology studies [5]. Secretome (20 µl) was separated by 4–12% Criterion™ XT Bis–Tris gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). Total protein was stained with MemCode™ Reversible Protein Stain Kit (Thermo Scientific #24580; Waltham, MA). Membranes were blocked for 1 h in 5% milk at room temperature and incubated with a primary antibody against Thbs1 (R&D Systems; #AF3074; 1:500) at 4 °C overnight. Recombinant Thbs1 (R&D Systems #7589-TH; 10 ng) was used as a positive control. Membranes were washed and incubated with secondary antibody (Vector #PI-1000; Malvern, PA, USA) at room temperature for 2 h. Images were detected using ECL Prime Western Blotting Detection Substrate (Amersham; Little Chalfont, UK). Protein expression was analyzed by densitometry using IQ-TL image analysis software (GE Healthcare; Waukesha, WI) and normalized to the total protein.
Angiogenesis assay
Mouse microvascular endothelial cells (Cell Biologics #C57-6024; Chicago, IL, USA; 2.5 × 104 cells/well, n = 2 technical replicates per condition) were seeded in 48-well plates coated with basement membrane extract (Matrigel® BD #354230; Franklin Lakes, NJ, USA). Cells were stimulated with either basal endothelial cell medium (CellBiologics #M1168), basal medium supplemented with 10% cardiac fibroblast secretome from the same samples used for RNA-Seq (n = 3 biological replicates), or endothelial growth medium (basal medium supplemented with VEGF, endothelial cell growth supplement, heparin, epidermal growth factor, hydrocortisone, l-glutamine, antibiotics, and FBS). Thbs1 was inhibited with a blocking antibody (10 µg/ml; Novus Biologicals #100-2059) as described previously [32]. As negative controls, endothelial cells were stimulated with the Thbs1 blocking antibody alone, an IgG1 isotype control (R&D #MAB002) alone, or basal medium diluted 1:10 with DMEM + 0.1% FBS. Images were acquired at 10X magnification following 22 h of stimulation. Angiogenesis variables were quantified using the ImageJ Angiogenesis Analyzer feature (http://image.bio.methods.free.fr/ImageJ/?Angiogenesis-Analyzer-for-ImageJ&artpage=2-6).
Statistics
Statistical analysis was performed according to the guidelines, Statistical Considerations in Reporting Cardiovascular Research [25]. All experiments were performed and analyzed in a blinded design. Data are presented as mean ± SEM unless otherwise noted. Survival rate was analyzed by Kaplan–Meier survival analysis. For physiology assessment, multiple group comparisons were made using one-way ANOVA followed by Newman–Keuls post hoc test. Statistics and bioinformatics for the RNA-seq are described above. RNA-seq comparisons to quantitative RT-PCR were made by Pearson’s linear regression analysis. Cell physiology comparisons were analyzed by two-tailed unpaired t test or one-sample t test. A value of p < 0.05 was considered statistically significant.
Results
Fibroblasts continually polarize over the MI spectrum
Proof of successful MI
Survival for the MI day 7 cohort was 52% (13/25), with 58% of mortality (7/12) due to cardiac rupture assessed at autopsy (Online Resource 1a). Infarct area (percent of LVI mass to total LV mass) did not differ between days 1, 3, and 7 (Online Resource 1b), indicating uniform infarct sizes across times and in line with the immediate myocyte necrosis that occurs after MI. As expected, MI led to infarct LV wall thinning, dilation, and decreased fractional shortening (Online Resource 1c–f). This is similar to what we and others have previously reported for mouse MI [9, 10, 21, 31, 66, 73].
MI differential gene expression and enrichment
Of 23,487 genes sequenced, 175 were duplicates and 6374 were removed either because all FPKM values were 0 or there were less than 3 replicates with values greater than 0 for any one group, leaving 16,938 genes for analysis (Fig. 1a; Online Table 1a). The FPKM values for all genes are displayed in Online Table 1. Of these, 3371 were differentially expressed by one-way ANOVA with Tukey’s post hoc test (Fig. 1a, b). Cell-specific markers for fibroblasts were highly expressed, while markers for other cell types were nearly undetectable (Online Resource 2a). Acta2 expression positively correlated with time after MI (r = 0.70, p = 0.01; Online Resource 2b). By mass spectrometry analysis of the day 0 and MI day 7 fibroblast secretomes, collagen I alpha 1 and 2 chains, secreted protein acidic and rich in cysteine (SPARC), and lysyl oxidase were significantly increased in the day 7 MI fibroblast secretome, while periostin showed no change (Online Resource 2b). Periostin gene expression was significantly increased in the LV infarct at days 1, 3, 5, and 7 (Online Resource 2c). Our results indicate the fibroblast activation marker collagens and collagen cross-linking enzymes are regulated at the protein level in MI cardiac fibroblasts.
a Workflow for analyzing differential gene expression. Of 23,487 genes sequenced, 175 were duplicates and 6,374 did not meet quality control criteria and were removed, leaving 16,938 for analysis. Of the remaining genes, 3,371 were differentially expressed by one-way ANOVA with Tukey’s post test (p < 0.05). b Heat map displaying differentially expressed genes across the MI time course. c Partial least-squares discriminant analysis (PLS-DA) plot. MI day 1 showed overlap with day 0, while days 3 and 7 were unique. d Enrichment analysis (Panther) for significant pathways across the MI time course. e Top 15 genes ranked by feature analysis and relative expression across each day (red—high; green—low)
For RT-PCR confirmation, four of the five genes (all except MMP-14) showed strong correlations between RNA-seq and RT-PCR (Online Resource 3). PLS-DA (Fig. 1c) indicated that each MI day was a separate cluster, with MI day 7 being the most separated from the day 0 no MI control. Significantly enriched pathways by Panther (all p < 0.05) are displayed in Fig. 1d. The top 15 genes ranked by important gene feature analysis are displayed in Fig. 1e, with the top-ranked gene being Thbs1.
By Venn diagram analysis of fold changes compared to day 0 with a fold-change cut-off of 1.2 and p < 0.05, (Fig. 2a), 387 genes were upregulated and 512 were downregulated at MI day 1, 553 were upregulated and 964 downregulated at MI day 3, and 672 upregulated and 868 downregulated at MI day 7. Very few of the top-ranked genes overlapped among days, with the majority being unique to only one time point (Fig. 2b). The top 5 genes at each MI day ranked by variable importance in projection coefficients are displayed in Fig. 2c.
MI yields distinct time-dependent gene expression profiles in cardiac fibroblasts. a Differentially expressed genes (upregulated or downregulated) at each MI day. b Venn diagrams indicating expression overlap among MI days. c Important feature analysis for the top five upregulated genes at each MI day
PLS-DA analysis and heat mapping for intracellular signaling genes (including FGF, IL-1, IL-4, MAPK, NFκB, PI3K, Smad, Socs, STAT, TGFβ1, and TNF) revealed that all MI days displayed unique signaling profiles compared to day 0, with MI days 1 and 3 showing the greatest overlap (Fig. 3a). By examining differentially expressed signaling genes in the heat map (Fig. 3b, c), day 0 fibroblasts use Pik3r4 and Stat5 signaling, while MI day 1 fibroblasts use Tnfrsf9 (CD137) signaling to induce inflammation [20], MI day 3 fibroblasts use interleukin-4 receptor alpha (Il4ra) signaling to stimulate anti-inflammatory and pro-fibrotic wound healing [44], and MI day 7 fibroblasts use Pik3r3 signaling to mediate TGFβ1 effects [65] and Fgfr2 to regulate PI3K signaling [42].
MI cardiac fibroblast signaling profiles. a Partial least-squares discriminant analysis (PLS-DA) for the most upregulated changes in signaling genes. b Heat map of differentially expressed signaling genes. c Gene expression (fold change over day 0) for differentially expressed signaling genes. n = 3 per group. *p < 0.05 versus day 0, #p < 0.05 versus MI day 1, and $p < 0.05 versus MI day 3
Cell-surface marker analysis
Using Enrichr, upregulated genes that encode for cell-surface proteins were compared across MI days to determine genes that distinguish fibroblasts and MI day. A total of 28 genes were selectively upregulated at MI day 1, 32 at MI day 3, and 31 at MI day 7. A total of 314 genes were all elevated with MI and did not change across MI time, and these can be considered potential pan MI fibroblast cell-surface markers. The top 25 ranked genes for each time are listed in Online Table 2. Recent mapping of the human cardiac proteome across cell types revealed a number of novel cardiac fibroblast cell-surface markers, including Acvr1, Met, Npr3, and Ror1 [11]. Our results indicate that Met and Npr3 were not affected by MI, while Acvr1 and Ror1 were both upregulated at MI day 7. These results provide a framework for identifying novel cardiac fibroblast cell-surface markers in the healthy and injured myocardium.
Cluster analysis of differentially expressed genes
We used Markov clustering of the gene correlation network to assess gene expression patterns over the MI time course. Differentially expressed genes fell into one of eight unique pattern clusters (Fig. 4a, b). Cluster 1 (276 genes) and 3 (156 genes) both represented genes upregulated only at day 1. The top enriched processes (Fig. 4) for these two clusters were pentose phosphate pathway and cytokine/chemokine-mediated inflammation, corroborating the results from Fig. 2a. Cluster 5 (88 genes) and 7 (49 genes) were increased at day 3 only, and were enriched for cholesterol biosynthesis (cluster 5) and cadherin signaling (cluster 7). Cluster 4 genes (109 genes) were upregulated at day 3 and further increased at day 7, and were enriched for angiogenesis. Cluster 2 genes (161 genes) were upregulated at day 7, and were enriched for cadherin signaling.
Distinct gene expression patterns by clustering. a Markov clustering analysis revealed eight distinct clusters of MI gene expression patterns. b Average fold change of genes in each cluster. c Number of genes and major enriched pathway for each gene cluster. n = 3 per group. *p < 0.05 versus day 0, #p < 0.05 versus MI day 1, and $p < 0.05 versus MI day 3
Day 0 fibroblasts were homeostatic
Cardiac fibroblasts in the uninjured heart were not quiescent. Rather, they served as sentinel cells to maintain the homeostasis of the myocardium by producing replacement ECM at low concentrations necessary for normal turnover [6]. We developed a list of pan fibroblast genes, which included genes that did not change across any of the times and ranked by FPKM value; the top 500 are shown in Online Table 3. Entering the top 500 ranked genes into the Mouse Gene Atlas (http://www.mouseatlas.org/) revealed close alignment with mouse embryonic fibroblasts and osteoblasts (Online Table 4). The major enriched pathways represent pathways that are highly active in fibroblasts, regardless of injury state, including integrin signaling and glycolysis. To characterize the day 0 phenotype, we analyzed genes that were high only at day 0 (genes significantly decreased at all MI days, n = 63 genes). By Panther analysis, day 0 genes were enriched for 5-hydroxytryptamine degradation, mannose metabolism, and de novo pyrimidine ribonucleotide biosynthesis (Online Table 5).
MI day 1 fibroblasts demonstrated a pro-inflammatory and leukocyte-recruiting, pro-survival, and anti-migratory signature (Fig. 5, Online Table 6)
Upregulated and downregulated genes are displayed by volcano plot (Fig. 5a). By Panther pathway analysis, chemokine- and cytokine-mediated inflammation was a major upregulated pathway. Interestingly, while Tgfb1 was increased at MI day 1 (Online Resource 4a), TGFβ signaling was downregulated (Fig. 5a). By GO pathway evaluation, regulation of T helper 17 cell lineage commitment (i.e. Il23a), positive regulation of memory T cell differentiation, negative regulation of interleukin-10 production, and positive regulation of macrophage chemotaxis (i.e., Ccl5, Csf1, and Cx3cl1) were upregulated biological processes (Online Table 6), indicating that MI day 1 fibroblasts provide leukocyte-recruiting signals to bring cells into the infarct region [68]. Out of ten apoptosis genes evaluated, only one, the anti-apoptotic Bcl2, was elevated at MI day 1 (Online Resource 4b), indicating pro-survival signaling. Fibroblast migration rates were significantly decreased at MI day 1 (Fig. 5b), and expression of migration-related genes Cthrc1, Fgf2, and Fzd2 was significantly decreased (Fig. 5c).
MI day 1 fibroblasts display a pro-inflammatory phenotype profile. a Volcano plot and enriched processes for upregulated and downregulated genes. b Fibroblast migration was significantly decreased at MI day 1 by one-sample t test. c Migration-related genes were significantly decreased at MI day 1 by unpaired t test. n = 3 per group. *p < 0.05 versus day 0
MI day 3 fibroblasts demonstrated a proliferative, pro-fibrotic, and pro-angiogenic signature
Upregulated and downregulated genes are plotted by volcano plot (Fig. 6a). By Panther pathway analysis (Fig. 6b), integrin signaling (i.e. Fn1 and Itga5), cadherin signaling (Cdh2), and angiogenesis (i.e., Vegfa) were major upregulated pathways (Online Table 5). By GO analysis, ECM organization (i.e., Fn1 and Lox) and cell migration involved in sprouting angiogenesis and blood vessel endothelial cell migration (i.e. Slit2, Ephb4, and Vegfa) were major upregulated biological processes (Online Table 6). Vegfa was significantly increased only at MI day 3. MI Day 3 fibroblasts were more proliferative than day 0 (Fig. 6c). The increase in fibroblast proliferation after MI is similar to what we and others have shown [13, 54]. Of the ten proliferation markers examined (Fig. 6d), the recently identified maker of fibroblast proliferation Ckap4 was elevated at MI day 3 [14]. In addition, expression of Casp3 was significantly reduced at MI day 3 (Online Resource 4b), indicating continuation of the anti-apoptotic and pro-survival signature. We observed robust changes in genes encoding for cholesterol biosynthesis (decreased at MI day 1 and increased at MI day 3). The increase in genes involved in cholesterol metabolism coincide with increased cell proliferation at MI day 3, which requires cholesterol biosynthesis to support new cell membrane formation, proper function of new cell membrane proteins, and lipid rafts/caveolae to mediate signaling pathways involved in cell proliferation [2, 51].
MI day 3 fibroblasts had a proliferative and pro-angiogenic phenotype profile. a Volcano plot and major upregulated pathways. b Vegfa expression was increased only at MI day 3. c Fibroblast proliferation by BrdU assay was increased by t test. d Of ten proliferation genes evaluated, Ckap4 increased at MI day 3. n = 3 per group. *p < 0.05 versus day 0, #p < 0.05 versus MI day 1, and $p < 0.05 versus MI day 3
MI day 7 fibroblasts demonstrated a homeostatic-like, anti-angiogenic signature
Upregulated and downregulated genes are plotted by volcano plot (Fig. 7a). By Panther pathway analysis (Online Table 5), cadherin signaling (i.e., Cdh11) and angiogenesis (i.e., Pdgfc, and Thbs1) were major upregulated pathways. By GO analysis (Online Table 6), positive regulation of bone mineralization (i.e., Grem1 and Tgfb3), wound healing, and regulation of endothelial cell chemotaxis (Sema5a, Tmsb4x, and Thbs1) were major upregulated processes. At MI day 7, the majority of fibroblasts are transdifferentiated into the myofibroblast phenotype [1]. Several myofibroblast-related genes were elevated only at MI day 7 (Online Resource 4a), including Cdh2 (N-cadherin) [48, 58], Cdh11 (cadherin-11) [53], Itga11 (integrin alpha-11) [1, 57], Timp1 and Timp3 [56], and Tmsb4x (thymosin beta-4) [4]. We also found that Wt1 and Twist1, markers of epicardial cell origin [52], were increased at day 7 (Online Resource 4c). Thbs1 was the highest ranked feature for differentially expressed genes by one-way ANOVA, being 3.2-fold increased at MI day 7 over day 0. By MI day 7, the fibroblast signature was moving towards a homeostatic-like phenotype, returning towards the new baseline established by the beginning formation of the infarct scar.
MI day 7 fibroblasts showed an anti-angiogenic myofibroblast phenotype profile. a Volcano plot and major upregulated processes. b Thbs1 expression was significantly increased; n = 3 per group. c LV infarct region from MI day 7 stained for cardiac fibroblast marker PDGFRα (green), Thbs1 (red), and DAPI (blue). Thbs1 was significantly increased within regions staining positive for PDGFRα; n = 4 day 0, n = 5 MI day 7. d Immunoblot analysis of Thbs1 in cardiac fibroblast secretome. MI significantly increased cardiac fibroblast secretion of Thbs1 at MI day 7; n = 3 per group; densitometry normalized to total membrane stain. *p < 0.05 versus day 0; $p < 0.05 versus MI day 3
As Thbs1 is a potent inhibitor of angiogenesis, we investigated cardiac fibroblast-derived Thbs1 as a potential factor regulating MI angiogenesis. Platelet-derived growth factor (PDGF) receptor alpha (PDGFRα)-positive cardiac fibroblasts demonstrated robust staining in the infarcted myocardium at MI day 7 relative to day 0 (Fig. 7c) consistent with other reports [15, 34, 35, 45]. Thbs1 immunostaining in cardiac fibroblasts (PDGFRα + cells) was significantly increased in day 7 infarcts (Fig. 7c). By immunoblotting, Thbs1 significantly increased in the secretome from isolated day 7 fibroblasts, indicating that MI day 7 fibroblasts actively secrete Thbs1 (Fig. 7d). CX3CL1 and CCL5 at MI day 1 and VEGF at MI day 3 did not localize to fibroblasts within the infarct, confirming that these proteins were secreted and not intracellular (Online Resource 6).
MI fibroblast dampening of angiogenesis is Thsb1-dependent
As angiogenic factors were differentially expressed after MI (Online Table 6), we investigated the effects of the MI fibroblast secretome on endothelial cell tube formation in vitro. The day 0 and MI day 3 secretomes stimulated angiogenesis, as assessed by increased total length, total branching length, total segment length, number of pieces, number of meshes, and number of junctions relative to basal media (Fig. 8). MI day 7 secretome decreased angiogenic variables relative to day 0, indicating that MI day 7 fibroblasts shut off angiogenesis. To determine whether Thbs1 explained the day 7 secretome effects on endothelial cells, Thbs1 was inhibited with a blocking antibody. Inhibition of Thbs1 reduced the effect of the day 7 secretome on angiogenesis variables, indicating that the inhibitory effect of the day 7 secretome on angiogenesis was Thbs1-dependent. Thbs1 inhibition did not alter the effects of the day 0 or MI day 3 secretome, and the Thbs1 blocking antibody alone control, IgG1 isotype antibody control, or the basal media diluted 1:10 with DMEM and 0.1% FBS media only control did not show any differences.
Effects of MI fibroblast secretome on in vitro endothelial cell tube formation. Day 0 and MI day 3 cardiac fibroblast secretome increased, while MI day 7 secretome decreased angiogenic variables relative to the effects of the day 0 secretome. MI day 7 secretome effects were inhibited by a Thbs1 blocking antibody. Scale bar = 200 µm. n = 3 per group. *p < 0.05 versus basal, #p < 0.05 versus day 0, $p < 0.05 versus MI day 7
Discussion
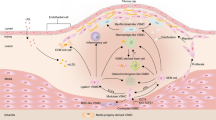
Cardiac fibroblasts have been implicated in MI remodeling and scar formation [15, 16, 33]. The goals of this study were to map the phenotypes of cardiac fibroblasts over the MI wound healing period and to explore new roles for fibroblasts in regulating inflammation and angiogenesis. The key findings were as follows: 1) day 0 no MI cardiac fibroblasts served as sentinel cells to actively maintain myocardial homeostasis; 2) cardiac fibroblasts underwent distinct transcriptomic changes at MI days 1, 3, and 7, with day 1 fibroblasts showing a pro-inflammatory signature, day 3 fibroblasts showing a proliferative, reparative, and pro-angiogenic profile, and day 7 fibroblasts showing homeostatic-like features and negatively regulating angiogenesis; and 3) the negative regulation of angiogenesis by day 7 fibroblasts occurred through Thbs1 signaling. Our results provide new insights into fibroblast phenotypes across the MI spectrum, with an emphasis on the roles of fibroblasts in regulating inflammation and angiogenesis (Fig. 9).
Identifying pan cell-surface markers for all cardiac fibroblasts has been a considerable challenge [6, 13, 14, 16, 17, 28]. Furthermore, a map of cell-surface markers across the MI injury state is lacking, as fibroblasts may increase or decrease marker expression in response to the changing microenvironment. Recent studies have identified novel markers in human cardiac fibroblasts from non-injured hearts, including the tyrosine kinase Ror1 and the activin receptor Acvr1 [11]. Our data validate these findings, as fibroblasts expressed both genes at every time point examined. The selective upregulation at MI day 7 indicates that these two genes may be particularly useful in identifying fibroblasts at MI day 7, when peak myofibroblast activation occurs [13]. Our findings provide a framework for identifying both novel constitutive and inducible markers.
In the absence of injury, the cardiac fibroblast is the major cell type producing and maintaining the ECM. Cardiac fibroblasts also support electro-chemical coupling, myocyte hypertrophy, and maintenance of the vasculature [17, 40]. Integrin signaling was the major enriched process for the pan fibroblast phenotype and was also enriched at MI days 3 and 7, indicating that MI fibroblasts selectively express integrins to adapt to the changing ECM environment. Integrin switching includes Itga5, the major fibronectin-binding integrin, at MI day 3, and Itga11, the major collagen-binding integrin, at MI day 7 [1]. Inflammation mediated by chemokine/cytokine signaling was enriched in the basal phenotype and at MI days 1 and 7, indicating that fibroblasts cross talk with resident immune cells regardless of injury state [29]. The day 0 secretome stimulated tube formation in vitro, indicating that fibroblasts in the uninjured heart play an active role in promoting a healthy vasculature.
At MI day 1, fibroblasts displayed an inflammatory signature, consistent with other reports that fibroblasts initiate and contribute to inflammation following injury [22, 38]. CCL5 and CX3CL1 are pro-inflammatory chemoattractants for monocytes and neutrophils [68]. Cardiac fibroblasts produce CX3CL1 in response to interferon γ stimulus [43]. Expression of Il23a, a pro-inflammatory cytokine that activates T helper 17 cells, was also increased at MI day 1 [67]. Colony stimulating facor-1 is critical for macrophage recruitment after MI [69]. TGFβ1 is an important anti-inflammatory cytokine that promotes fibroblast activation to myofibroblasts. TGFβ signaling was a major downregulated process, indicating that MI day 1 fibroblasts are unresponsive to increased TGFβ1 expression. Our results indicate that, in addition to other cell sources, fibroblasts are a source of pro-inflammatory chemokines and cytokines to recruit inflammatory cells to the infarct region. Bcl2 was highly increased at MI day 1, indicating that fibroblasts respond to ischemia by promoting cell survival signaling [13].
At MI day 3, fibroblasts showed a proliferative, pro-fibrotic, and pro-angiogenic profile. Cholesterol biosynthesis, a major upregulated process at day 3, is important in supporting cardiac fibroblast proliferation. Treatment of mice with simvastatin, a potent inhibitor of cholesterol biosynthesis, decreases LV collagen deposition and fibroblast activity after MI [55], and in vitro, simvastatin induces cardiac fibroblast apoptosis, with a stronger effect on quiescent fibroblasts than activated myofibroblasts [8]. In line with these results, cholesterol biosynthesis was elevated at MI day 3, at which point fibroblasts were highly proliferative [13]. Thus, cholesterol biosynthesis may be required for MI fibroblast proliferation during the early granulation phase prior to scar formation and maturation.
MI day 3 fibroblasts showed a pro-angiogenic profile. Cardiac fibroblasts secrete several factors that regulate angiogenesis, including VEGF [40, 60, 71, 72]. The positive regulation of angiogenesis at MI day 3 coincides with the peak of vessel formation and endothelial cell proliferation in the mouse, indicating that fibroblasts are a significant in vivo contributor to endothelial cell activation [63].
At MI day 7, fibroblasts displayed a homeostatic-like, anti-angiogenic signature. The epicardial cell markers Wt1 and Twist1 were significantly increased, consistent with other reports, indicating that MI day 7 fibroblasts are derived from epicardial progenitors that underwent epithelial-to-mesenchymal transition [13, 47, 52]. In contrast to MI day 3, day 7 fibroblasts showed an anti-angiogenic profile. Fibroblast-derived factors can inhibit angiogenesis [30]. Our results suggest that day 7 fibroblasts may inhibit angiogenesis by altering the balance between pro-angiogenic (Vegfa) and anti-angiogenic (Cxcl14, Sema3a, Lypd1, and Thbs1) soluble mediators. Thbs1 has been heavily implicated as a potent inhibitor of angiogenesis, mainly through pro-apoptotic and anti-VEGF actions on cardiac microvascular endothelial cells [71]. Thbs1 expression is induced in the infarcted heart by macrophages, platelets, and fibroblasts, peaking 24 h and remaining elevated as long as 28 days after MI in the rat heart [12, 50, 70]. Mice lacking Thbs1 show a prolonged inflammatory response and expansion of granulation tissue following MI [12]. Our results indicate that cardiac fibroblasts secrete Thbs1 at day 7 but not day 3, and that blocking Thbs1 reverses the anti-angiogenic properties of the MI day 7 secretome, indicating that fibroblast-derived Thbs1 tempers excessive angiogenesis.
Fibroblasts from day 0 healthy hearts potently stimulated angiogenesis, indicating that rather than being quiescent, fibroblasts in the healthy heart actively support angiogenesis and vessel homeostasis. Our results indicate a dichotomous role in fibroblast regulation of angiogenesis following MI injury, as day 0 and day 3 fibroblasts promoted angiogenesis, while day 7 fibroblasts show decreased angiogenic potential. Angiogenesis is typically considered beneficial for MI remodeling and correlates with better outcomes [31], thus targeting cardiac fibroblasts could be a potential therapeutic strategy to improve MI angiogenesis. MI day 7 fibroblasts may temper excessive and uncontrolled angiogenesis that could otherwise perturb proper scar formation [49]. Anti-inflammatory and pro-reparative resident macrophage and circulating monocyte populations promote angiogenesis in the MI heart. It is possible that macrophages are the predominant cell types in the MI heart that promote angiogenesis, while fibroblasts dial up or down as needed [39]. In addition to inhibiting angiogenesis, Thbs1 promotes the resolution of inflammation, suggesting that anti-inflammatory signals such as TGFβ1 may induce Thbs1 expression and promote anti-angiogenic actions during scar maturation [12].
Our results reveal that some concepts of classical fibroblast activation are in play after MI, while others have been preconceived ideas. Of note, we did not observe the expected increase in the myofibroblast marker Postn in MI fibroblasts. Periostin gene or protein expression did not change after MI in isolated fibroblasts, while gene expression did increase in the LV infarct at days 1, 3, 5, and 7. A recent report has shown increased Postn in the Tcf21+ subpopulation of cardiac cells examined [13], indicating that a subset of fibroblasts (Tcf21+ cells) and non-fibroblast cells (e.g., smooth muscle cells and macrophages) are the source of periostin. We recently reported on the MI macrophage polarization time course [36]. Tcf21 was expressed in no MI resident cardiac macrophages at 25–50-fold higher levels than MI days 1, 3, or 7 macrophages and lower expression than fibroblasts (all p < 0.01), revealing fluidity in cell marker expression across cell types (fibroblasts and macrophages) and across MI times. We evaluated the total fibroblast pool (all fibroblasts adhering to culture dishes), indicating that periostin is a selective marker for a subset of cells. Despite culturing, our cells retained the features of myofibroblast activation including increased collagen, SPARC, and lysyl oxidase secretion. Our results reveal that, rather than a binary on or off pattern, Acta2 gene expression actually shows a continual increase over time (linear regression correlation coefficient r = 0.70, p = 0.01), indicating a persistent shift in time to the myofibroblast phenotype. Based on our evaluation, fibroblasts retained the in vivo MI phenotype in vitro through passage 3.
In conclusion, cardiac fibroblasts show distinct transcriptomic profiles at different time points over the first week of MI wound healing. Our findings provide novel insights into the potential mechanisms and pathways that regulate fibroblast physiology during the wound healing phases of cardiac remodeling. Overall, our work suggests that attention to temporal profiles should be given when considering interventions that target fibroblasts.
References
Bansal R, Nakagawa S, Yazdani S, van Baarlen J, Venkatesh A, Koh AP, Song WM, Goossens N, Watanabe H, Beasley MB, Powell CA, Storm G, Kaminski N, van Goor H, Friedman SL, Hoshida Y, Prakash J (2017) Integrin alpha 11 in the regulation of the myofibroblast phenotype: implications for fibrotic diseases. Exp Mol Med 49:e396. https://doi.org/10.1038/emm.2017.213
Beloribi-Djefaflia S, Vasseur S, Guillaumond F (2016) Lipid metabolic reprogramming in cancer cells. Oncogenesis 5:e189. https://doi.org/10.1038/oncsis.2015.49
Besnier M, Galaup A, Nicol L, Henry JP, Coquerel D, Gueret A, Mulder P, Brakenhielm E, Thuillez C, Germain S, Richard V, Ouvrard-Pascaud A (2014) Enhanced angiogenesis and increased cardiac perfusion after myocardial infarction in protein tyrosine phosphatase 1B-deficient mice. FASEB J 28:3351–3361. https://doi.org/10.1096/fj.13-245753
Bollini S, Riley PR, Smart N (2015) Thymosin beta4: multiple functions in protection, repair and regeneration of the mammalian heart. Expert Opin Biol Ther 15(Suppl 1):S163–S174. https://doi.org/10.1517/14712598.2015.1022526
Brooks HL, Lindsey ML (2018) Guidelines for authors and reviewers on antibody use in physiology studies. Am J Physiol Heart Circul Physiol 314:H724–H732. https://doi.org/10.1152/ajpheart.00512.2017
Camelliti P, Borg TK, Kohl P (2005) Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res 65:40–51. https://doi.org/10.1016/j.cardiores.2004.08.020
Cochain C, Channon KM, Silvestre JS (2013) Angiogenesis in the infarcted myocardium. Antioxid Redox Signal 18:1100–1113. https://doi.org/10.1089/ars.2012.4849
Copaja M, Venegas D, Aranguiz P, Canales J, Vivar R, Catalan M, Olmedo I, Rodriguez AE, Chiong M, Leyton L, Lavandero S, Diaz-Araya G (2011) Simvastatin induces apoptosis by a Rho-dependent mechanism in cultured cardiac fibroblasts and myofibroblasts. Toxicol Appl Pharmacol 255:57–64. https://doi.org/10.1016/j.taap.2011.05.016
DeLeon-Pennell KY, de Castro Bras LE, Iyer RP, Bratton DR, Jin YF, Ripplinger CM, Lindsey ML (2014) P. gingivalis lipopolysaccharide intensifies inflammation post-myocardial infarction through matrix metalloproteinase-9. J Mol Cell Cardiol 76:218–226. https://doi.org/10.1016/j.yjmcc.2014.09.007
DeLeon-Pennell KY, Iyer RP, Ero OK, Cates CA, Flynn ER, Cannon PL, Jung M, Shannon D, Garrett MR, Buchanan W, Hall ME, Ma Y, Lindsey ML (2017) Periodontal-induced chronic inflammation triggers macrophage secretion of Ccl12 to inhibit fibroblast-mediated cardiac wound healing. JCI Insight. https://doi.org/10.1172/jci.insight.94207
Doll S, Dressen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch MA, Lahm H, Lange R, Krane M, Mann M (2017) Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun 8:1469. https://doi.org/10.1038/s41467-017-01747-2
Frangogiannis NG, Ren G, Dewald O, Zymek P, Haudek S, Koerting A, Winkelmann K, Michael LH, Lawler J, Entman ML (2005) Critical role of endogenous thrombospondin-1 in preventing expansion of healing myocardial infarcts. Circulation 111:2935–2942. https://doi.org/10.1161/CIRCULATIONAHA.104.510354
Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, Molkentin JD (2018) Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 128:2127–2143. https://doi.org/10.1172/JCI98215
Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E (2018) Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 138:166–180. https://doi.org/10.1161/CIRCULATIONAHA.117.030742
He L, Huang X, Kanisicak O, Li Y, Wang Y, Li Y, Pu W, Liu Q, Zhang H, Tian X, Zhao H, Liu X, Zhang S, Nie Y, Hu S, Miao X, Wang QD, Wang F, Chen T, Xu Q, Lui KO, Molkentin JD, Zhou B (2017) Preexisting endothelial cells mediate cardiac neovascularization after injury. J Clin Invest 127:2968–2981. https://doi.org/10.1172/JCI93868
Ivey MJ, Kuwabara JT, Pai JT, Moore RE, Sun Z, Tallquist MD (2018) Resident fibroblast expansion during cardiac growth and remodeling. J Mol Cell Cardiol 114:161–174. https://doi.org/10.1016/j.yjmcc.2017.11.012
Ivey MJ, Tallquist MD (2016) Defining the Cardiac Fibroblast. Circ J 80:2269–2276. https://doi.org/10.1253/circj.CJ-16-1003
Iyer RP, de Castro Bras LE, Cannon PL, Ma Y, DeLeon-Pennell KY, Jung M, Flynn ER, Henry JB, Bratton DR, White JA, Fulton LK, Grady AW, Lindsey ML (2016) Defining the sham environment for post-myocardial infarction studies in mice. Am J Physiol Heart Circul Physiol 311:H822–H836. https://doi.org/10.1152/ajpheart.00067.2016
Iyer RP, Patterson NL, Zouein FA, Ma Y, Dive V, de Castro Bras LE, Lindsey ML (2015) Early matrix metalloproteinase-12 inhibition worsens post-myocardial infarction cardiac dysfunction by delaying inflammation resolution. Int J Cardiol 185:198–208. https://doi.org/10.1016/j.ijcard.2015.03.054
Jung IH, Oh GT (2016) The roles of CD137 signaling in atherosclerosis. Korean Circ J 46:753–761. https://doi.org/10.4070/kcj.2016.46.6.753
Jung M, Ma Y, Iyer RP, DeLeon-Pennell KY, Yabluchanskiy A, Garrett MR, Lindsey ML (2017) IL-10 improves cardiac remodeling after myocardial infarction by stimulating M2 macrophage polarization and fibroblast activation. Basic Res Cardiol 112:33. https://doi.org/10.1007/s00395-017-0622-5
Kawaguchi M, Takahashi M, Hata T, Kashima Y, Usui F, Morimoto H, Izawa A, Takahashi Y, Masumoto J, Koyama J, Hongo M, Noda T, Nakayama J, Sagara J, Taniguchi S, Ikeda U (2011) Inflammasome activation of cardiac fibroblasts is essential for myocardial ischemia/reperfusion injury. Circulation 123:594–604. https://doi.org/10.1161/CIRCULATIONAHA.110.982777
Lindsey ML (2018) Assigning matrix metalloproteinase roles in ischaemic cardiac remodelling. Nat Rev Cardiol 15:471–479. https://doi.org/10.1038/s41569-018-0022-z
Lindsey ML, Bolli R, Canty JM, Du XJ, Frangogiannis NG, Frantz S, Gourdie RG, Holmes JW, Jones SP, Kloner R, Lefer DJ, Liao R, Murphy E, Ping P, Przyklenk K, Recchia FA, Schwartz Longacre L, Ripplinger CM, Van Eyk JE, Heusch G (2018) Guidelines for experimental models of myocardial ischemia and infarction. Am J Physiol Heart Circul Physiol 314:H812–H838. https://doi.org/10.1152/ajpheart.00335.2017
Lindsey ML, Gray GA, Wood SK, Curran-Everett D (2018) Statistical considerations in reporting cardiovascular research. Am J Physiol Heart Circul Physiol 315:H303–H313. https://doi.org/10.1152/ajpheart.00309.2018
Lindsey ML, Iyer RP, Zamilpa R, Yabluchanskiy A, DeLeon-Pennell KY, Hall ME, Kaplan A, Zouein FA, Bratton D, Flynn ER, Cannon PL, Tian Y, Jin YF, Lange RA, Tokmina-Roszyk D, Fields GB, de Castro Bras LE (2015) A novel collagen matricryptin reduces left ventricular dilation post-myocardial infarction by promoting scar formation and angiogenesis. J Am Coll Cardiol 66:1364–1374. https://doi.org/10.1016/j.jacc.2015.07.035
Lindsey ML, Kassiri Z, Virag JAI, de Castro Bras LE, Scherrer-Crosbie M (2018) Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circul Physiol 314:H733–H752. https://doi.org/10.1152/ajpheart.00339.2017
Ma Y, Iyer RP, Jung M, Czubryt MP, Lindsey ML (2017) Cardiac fibroblast activation post-myocardial infarction: current knowledge gaps. Trends Pharmacol Sci 38:448–458. https://doi.org/10.1016/j.tips.2017.03.001
Ma Y, Mouton AJ, Lindsey ML (2018) Cardiac macrophage biology in the steady-state heart, the aging heart, and following myocardial infarction. Transl Res 191:15–28. https://doi.org/10.1016/j.trsl.2017.10.001
Masuda S, Matsuura K, Shimizu T (2018) Inhibition of LYPD1 is critical for endothelial network formation in bioengineered tissue with human cardiac fibroblasts. Biomaterials 166:109–121. https://doi.org/10.1016/j.biomaterials.2018.03.002
Meschiari CA, Jung M, Iyer RP, Yabluchanskiy A, Toba H, Garrett MR, Lindsey ML (2018) Macrophage overexpression of matrix metalloproteinase-9 in aged mice improves diastolic physiology and cardiac wound healing after myocardial infarction. Am J Physiol Heart Circul Physiol 314:H224–H235. https://doi.org/10.1152/ajpheart.00453.2017
Michaud-Levesque J, Richard S (2009) Thrombospondin-1 is a transcriptional repression target of PRMT6. J Biol Chem 284:21338–21346. https://doi.org/10.1074/jbc.M109.005322
Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, Gunaje J, Otsu K, Davis J (2017) Fibroblast-specific genetic manipulation of p38 mitogen-activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation 136:549–561. https://doi.org/10.1161/CIRCULATIONAHA.116.026238
Moore-Morris T, Cattaneo P, Guimaraes-Camboa N, Bogomolovas J, Cedenilla M, Banerjee I, Ricote M, Kisseleva T, Zhang L, Gu Y, Dalton ND, Peterson KL, Chen J, Puceat M, Evans SM (2018) Infarct fibroblasts do not derive from bone marrow lineages. Circ Res 122:583–590. https://doi.org/10.1161/CIRCRESAHA.117.311490
Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, Stallcup WB, Gu Y, Dalton ND, Cedenilla M, Gomez-Amaro R, Zhou B, Brenner DA, Peterson KL, Chen J, Evans SM (2014) Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest 124:2921–2934. https://doi.org/10.1172/JCI74783
Mouton AJ, DeLeon-Pennell KY, Rivera Gonzalez OJ, Flynn ER, Freeman TC, Saucerman JJ, Garrett MR, Ma Y, Harmancey R, Lindsey ML (2018) Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol 113:26. https://doi.org/10.1007/s00395-018-0686-x
Munoz-Rodriguez C, Fernandez S, Osorio JM, Olivares F, Anfossi R, Bolivar S, Humeres C, Boza P, Vivar R, Pardo-Jimenez V, Hemmings KE, Turner NA, Diaz-Araya G (2018) Expression and function of TLR4-induced B1R bradykinin receptor on cardiac fibroblasts. Toxicol Appl Pharmacol 351:46–56. https://doi.org/10.1016/j.taap.2018.05.011
Nagaraju CK, Dries E, Popovic N, Singh AA, Haemers P, Roderick HL, Claus P, Sipido KR, Driesen RB (2017) Global fibroblast activation throughout the left ventricle but localized fibrosis after myocardial infarction. Sci Rep 7:10801. https://doi.org/10.1038/s41598-017-09790-1
Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ (2007) The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 204:3037–3047. https://doi.org/10.1084/jem.20070885
Nehls V, Herrmann R, Huhnken M, Palmetshofer A (1998) Contact-dependent inhibition of angiogenesis by cardiac fibroblasts in three-dimensional fibrin gels in vitro: implications for microvascular network remodeling and coronary collateral formation. Cell Tissue Res 293:479–488
Nielsen SH, Mouton AJ, DeLeon-Pennell KY, Genovese F, Karsdal M, Lindsey ML (2017) Understanding cardiac extracellular matrix remodeling to develop biomarkers of myocardial infarction outcomes. Matrix Biol. https://doi.org/10.1016/j.matbio.2017.12.001
Ornitz DM, Itoh N (2015) The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4:215–266. https://doi.org/10.1002/wdev.176
Pappritz K, Savvatis K, Koschel A, Miteva K, Tschope C, Van Linthout S (2018) Cardiac (myo)fibroblasts modulate the migration of monocyte subsets. Sci Rep 8:5575. https://doi.org/10.1038/s41598-018-23881-7
Peng H, Sarwar Z, Yang XP, Peterson EL, Xu J, Janic B, Rhaleb N, Carretero OA, Rhaleb NE (2015) Profibrotic role for interleukin-4 in cardiac remodeling and dysfunction. Hypertension 66:582–589. https://doi.org/10.1161/HYPERTENSIONAHA.115.05627
Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, Tallquist MD (2016) Revisiting Cardiac Cellular Composition. Circ Res 118:400–409. https://doi.org/10.1161/CIRCRESAHA.115.307778
Prescott MJ, Lidster K (2017) Improving quality of science through better animal welfare: the NC3Rs strategy. Lab Anim (NY) 46:152–156. https://doi.org/10.1038/laban.1217
Ruiz-Villalba A, Simon AM, Pogontke C, Castillo MI, Abizanda G, Pelacho B, Sanchez-Dominguez R, Segovia JC, Prosper F, Perez-Pomares JM (2015) Interacting resident epicardium-derived fibroblasts and recruited bone marrow cells form myocardial infarction scar. J Am Coll Cardiol 65:2057–2066. https://doi.org/10.1016/j.jacc.2015.03.520
Schroer AK, Merryman WD (2015) Mechanobiology of myofibroblast adhesion in fibrotic cardiac disease. J Cell Sci 128:1865–1875. https://doi.org/10.1242/jcs.162891
Schwarz ER, Speakman MT, Patterson M, Hale SS, Isner JM, Kedes LH, Kloner RA (2000) Evaluation of the effects of intramyocardial injection of DNA expressing vascular endothelial growth factor (VEGF) in a myocardial infarction model in the rat–angiogenesis and angioma formation. J Am Coll Cardiol 35:1323–1330
Sezaki S, Hirohata S, Iwabu A, Nakamura K, Toeda K, Miyoshi T, Yamawaki H, Demircan K, Kusachi S, Shiratori Y, Ninomiya Y (2005) Thrombospondin-1 is induced in rat myocardial infarction and its induction is accelerated by ischemia/reperfusion. Exp Biol Med (Maywood) 230:621–630
Singh P, Saxena R, Srinivas G, Pande G, Chattopadhyay A (2013) Cholesterol biosynthesis and homeostasis in regulation of the cell cycle. PLoS One 8:e58833. https://doi.org/10.1371/journal.pone.0058833
Smits AM, Dronkers E, Goumans MJ (2018) The epicardium as a source of multipotent adult cardiac progenitor cells: their origin, role and fate. Pharmacol Res 127:129–140. https://doi.org/10.1016/j.phrs.2017.07.020
Souders CA, Bowers SL, Baudino TA (2009) Cardiac fibroblast: the renaissance cell. Circ Res 105:1164–1176. https://doi.org/10.1161/CIRCRESAHA.109.209809
Squires CE, Escobar GP, Payne JF, Leonardi RA, Goshorn DK, Sheats NJ, Mains IM, Mingoia JT, Flack EC, Lindsey ML (2005) Altered fibroblast function following myocardial infarction. J Mol Cell Cardiol 39:699–707. https://doi.org/10.1016/j.yjmcc.2005.07.008
Sun F, Duan W, Zhang Y, Zhang L, Qile M, Liu Z, Qiu F, Zhao D, Lu Y, Chu W (2015) Simvastatin alleviates cardiac fibrosis induced by infarction via up-regulation of TGF-beta receptor III expression. Br J Pharmacol 172:3779–3792. https://doi.org/10.1111/bph.13166
Takawale A, Zhang P, Patel VB, Wang X, Oudit G, Kassiri Z (2017) Tissue inhibitor of matrix metalloproteinase-1 promotes myocardial fibrosis by mediating CD63-integrin beta1 interaction. Hypertension 69:1092–1103. https://doi.org/10.1161/HYPERTENSIONAHA.117.09045
Talior-Volodarsky I, Connelly KA, Arora PD, Gullberg D, McCulloch CA (2012) alpha11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovasc Res 96:265–275. https://doi.org/10.1093/cvr/cvs259
Thompson SA, Blazeski A, Copeland CR, Cohen DM, Chen CS, Reich DM, Tung L (2014) Acute slowing of cardiac conduction in response to myofibroblast coupling to cardiomyocytes through N-cadherin. J Mol Cell Cardiol 68:29–37. https://doi.org/10.1016/j.yjmcc.2013.12.025
Tomita K, Takashina M, Mizuno N, Sakata K, Hattori K, Imura J, Ohashi W, Hattori Y (2015) Cardiac fibroblasts: contributory role in septic cardiac dysfunction. J Surg Res 193:874–887. https://doi.org/10.1016/j.jss.2014.09.012
Twardowski RL, Black LD 3rd (2014) Cardiac fibroblasts support endothelial cell proliferation and sprout formation but not the development of multicellular sprouts in a fibrin gel co-culture model. Ann Biomed Eng 42:1074–1084. https://doi.org/10.1007/s10439-014-0971-2
van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J (2010) Myocardial remodeling after infarction: the role of myofibroblasts. Nat Rev Cardiol 7:30–37. https://doi.org/10.1038/nrcardio.2009.199
van Nieuwenhoven FA, Hemmings KE, Porter KE, Turner NA (2013) Combined effects of interleukin-1alpha and transforming growth factor-beta1 on modulation of human cardiac fibroblast function. Matrix Biol 32:399–406. https://doi.org/10.1016/j.matbio.2013.03.008
Virag JI, Murry CE (2003) Myofibroblast and endothelial cell proliferation during murine myocardial infarct repair. Am J Pathol 163:2433–2440. https://doi.org/10.1016/S0002-9440(10)63598-5
Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:11033. https://doi.org/10.1093/nar/gkw880
Voloshenyuk TG, Landesman ES, Khoutorova E, Hart AD, Gardner JD (2011) Induction of cardiac fibroblast lysyl oxidase by TGF-beta1 requires PI3 K/Akt, Smad3, and MAPK signaling. Cytokine 55:90–97. https://doi.org/10.1016/j.cyto.2011.03.024
Weirather J, Hofmann UD, Beyersdorf N, Ramos GC, Vogel B, Frey A, Ertl G, Kerkau T, Frantz S (2014) Foxp3 + CD4+ T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ Res 115:55–67. https://doi.org/10.1161/CIRCRESAHA.115.303895
Wu L, Diny NL, Ong S, Barin JG, Hou X, Rose NR, Talor MV, Cihakova D (2016) Pathogenic IL-23 signaling is required to initiate GM-CSF-driven autoimmune myocarditis in mice. Eur J Immunol 46:582–592. https://doi.org/10.1002/eji.201545924
Yabluchanskiy A, Ma Y, DeLeon-Pennell KY, Altara R, Halade GV, Voorhees AP, Nguyen NT, Jin YF, Winniford MD, Hall ME, Han HC, Lindsey ML (2016) Myocardial infarction superimposed on aging: mMP-9 Deletion promotes M2 macrophage polarization. J Gerontol A Biol Sci Med Sci 71:475–483. https://doi.org/10.1093/gerona/glv034
Yano T, Miura T, Whittaker P, Miki T, Sakamoto J, Nakamura Y, Ichikawa Y, Ikeda Y, Kobayashi H, Ohori K, Shimamoto K (2006) Macrophage colony-stimulating factor treatment after myocardial infarction attenuates left ventricular dysfunction by accelerating infarct repair. J Am Coll Cardiol 47:626–634. https://doi.org/10.1016/j.jacc.2005.09.037
Zhao C, Isenberg JS, Popel AS (2018) Human expression patterns: qualitative and quantitative analysis of thrombospondin-1 under physiological and pathological conditions. J Cell Mol Med 22:2086–2097. https://doi.org/10.1111/jcmm.13565
Zhao L, Eghbali-Webb M (2001) Release of pro- and anti-angiogenic factors by human cardiac fibroblasts: effects on DNA synthesis and protection under hypoxia in human endothelial cells. Biochem Biophys Acta 1538:273–282
Zhao W, Zhao T, Huang V, Chen Y, Ahokas RA, Sun Y (2011) Platelet-derived growth factor involvement in myocardial remodeling following infarction. J Mol Cell Cardiol 51:830–838. https://doi.org/10.1016/j.yjmcc.2011.06.023
Zhu M, Goetsch SC, Wang Z, Luo R, Hill JA, Schneider J, Morris SM Jr, Liu ZP (2015) FoxO4 promotes early inflammatory response upon myocardial infarction via endothelial Arg1. Circ Res 117:967–977. https://doi.org/10.1161/CIRCRESAHA.115.306919
Acknowledgements
We thank Dr. Rugmani P. Iyer, Dr. Mira Jung, and Presley C. Cannon for their technical support. We acknowledge funding from the American Heart Association under Award Numbers 15SDG22930009 and 18POST34000039, from the National Institutes of Health under Award Numbers GM103328, GM103476, GM104357, GM114833, GM115428, HL051971, HL075360, HL105324, and HL129823, and from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development under Award Numbers 5I01BX000505 and IK2BX003922. T.C.F. is funded by an Institute Strategic Programme Grant funding from the Biotechnology and Biological Sciences Research Council (BB/J004227/1). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies. All authors have reviewed and approved the article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have read the journal authorship agreement and policy on disclosure of potential conflicts of interest and have nothing to disclose.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mouton, A.J., Ma, Y., Rivera Gonzalez, O.J. et al. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Res Cardiol 114, 6 (2019). https://doi.org/10.1007/s00395-019-0715-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00395-019-0715-4