Abstract
Purpose
Recently, microRNA-124 (miR-124) has been demonstrated as a potential tumor suppressor in several types of cancers. However, the role of miR-124 in colorectal cancer remains unclear. This study was aimed at investigating the clinicopathological significance of miR-124 expression in colorectal cancer.
Methods
Quantitative real-time PCR was used to analyze miR-124 expression in 96 colorectal cancers and individual-matched normal mucosa samples. The expression of miR-124 was assessed for associations with clinicopathological characteristics using chi-square test. The survival curves were calculated by the Kaplan–Meier method. The influence of each variable on survival was examined by the Cox multivariate regression analysis.
Results
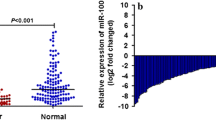
The miR-124 expression was significantly downregulated in colorectal cancer compared to normal mucosa (P = 0.001). In colorectal cancer, miR-124 decreased expression correlated significantly with the grade of differentiation (P = 0.021). Univariate survival analysis showed that the downregulated miR-124 was significantly correlated with worse prognosis, both in terms of overall survival (P = 0.017) and disease-free survival (DFS) (P = 0.014). Further, the downregulated miR-124 was demonstrated as a prognostic factor for overall survival (hazard ratio, HR = 4.634; 95 % confidence interval, CI, 1.731–12.404; P = 0.002) and DFS (HR = 4.533, 95 % CI 1.733–11.856, P = 0.002), independently of gender, age, location, maximum tumor size, depth of invasion, differentiation, and TNM stage.
Conclusions
MiR-124 may play a certain role in the development of colorectal cancer. The downregulation expression of miR-124 is an independent prognostic factor in patients with colorectal cancer.


Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Calin GA, Dumitru CD, Shimizu M et al (2002) Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99:15524–15529
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Schetter AJ, Leung SY, Sohn JJ et al (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299:425–436
Aaltonen LA et al (2000) World Health Organization classification of tumors. Pathology and genetics of tumours of the digestive system. International Agency for Research on Cancer Press, Lyon
American Cancer Society (2008) Colorectal cancer facts & figures 2008–2010. American Cancer Society, Atlanta
Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ (2003) Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1:882–891
Nugent M, Miller N, Kerin MJ (2011) MicroRNAs in colorectal cancer: function, dysregulation and potential as novel biomarkers. Eur J Surg Oncol 37:649–654
Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ (2011) MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis 32:247–253
Pierson J, Hostager B, Fan R, Vibhakar R (2008) Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 90:1–7
Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD (2010) Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 9:167
Patnaik SK, Kannisto E, Knudsen S, Yendamuri S (2010) Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res 70:36–45
Hatziapostolou M, Polytarchou C, Aggelidou E (2011) An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147:1233–1247
Hunt S, Jones AV, Hinsley EE, Whawell SA, Lambert DW (2011) MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett 585:187–192
Zheng F, Liao YJ, Cai MY et al (2012) The putative tumour suppressor microRNA-12 4 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. GUT 61:278–289
Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T (2009) DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer 124:2367–2374
Lv XB, Jiao Y, Qing Y, Hu H, Cui X, Lin T, Song E, Yu F (2011) miR-124 suppresses multiple steps of breast cancer metastasis by targeting a cohort of pro-metastatic genes in vitro. Chin J Cancer 30:821–830
Chen C, Ridzon DA, Broomer AJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179
Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF (2009) Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers 26:27–34
Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J (2010) miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31:766–776
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Method 25:402–408
Ponomarev ED, Veremeyko T, Barteneva NS (2011) Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC Res Notes 4:517
Suzuki H, Takatsuka S, Akashi H, Yamamoto E, Nojima M, Maruyama R, Kai M, Yamano HO, Sasaki Y, Tokino T, Shinomura Y, Imai K, Toyota M (2011) Genome-wide profiling of chromatin signatures reveals epigenetic regulation of microRNA genes in colorectal cancer. Cancer Res 71:5646–5658
Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J (2006) Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 5:29
Lujambio A, Ropero S, Ballestar E et al (2007) Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67:1424–1429
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455:64–71
Suzuki H, Igarashi S, Nojima M et al (2010) IGFBP7 is a p53 responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 31:342–349
Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132:363–374
Silber J, Lim DA, Petritsch C et al (2008) miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6:14
Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448
Grossel MJ, Hinds PW (2006) From cell cycle to differentiation: an expanding role for cdk6. Cell Cycle 5:266–270
Huang TC, Chang HY, Chen CY, Wu PY, Lee H, Liao YF, Hsu WM, Huang HC, Juan HF (2011) Silencing of miR-124 induces neuroblastoma SK-N-SH cell differentiation, cell cycle arrest and apoptosis through promoting AHR. FEBS Lett 585:3582–3586
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 30830100) and Ph.D. Programs Foundation of the Ministry of Education of China (no. 200806100058). We thank Chen Keling and Lv Zhaoying for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, MJ., Li, Y., Wang, R. et al. Downregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancer. Int J Colorectal Dis 28, 183–189 (2013). https://doi.org/10.1007/s00384-012-1550-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00384-012-1550-3