Abstract
Objectives
We tried to realize accurate pathological classification, assessment of prognosis, and genomic molecular typing of renal cell carcinoma by CT texture feature analysis. To determine whether CT texture features can perform accurate pathological classification and evaluation of prognosis and genomic characteristics in renal cell carcinoma.
Methods
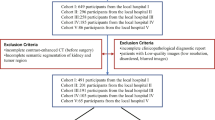
Patients with renal cell carcinoma from five open-source cohorts were analyzed retrospectively in this study. These data were randomly split to train and test machine learning algorithms to segment the lesion, predict the histological subtype, tumor stage, and pathological grade. Dice coefficient and performance metrics such as accuracy and AUC were calculated to evaluate the segmentation and classification model. Quantitative decomposition of the predictive model was conducted to explore the contribution of each feature. Besides, survival analysis and the statistical correlation between CT texture features, pathological, and genomic signatures were investigated.
Results
A total of 569 enhanced CT images of 443 patients (mean age 59.4, 278 males) were included in the analysis. In the segmentation task, the mean dice coefficient was 0.96 for the kidney and 0.88 for the cancer region. For classification of histologic subtype, tumor stage, and pathological grade, the model was on a par with radiologists and the AUC was 0.83 \(\pm\) 0.1, 0.80 \(\pm\) 0.1, and 0.77 \(\pm\) 0.1 at 95% confidence intervals, respectively. Moreover, specific quantitative CT features related to clinical prognosis were identified. A strong statistical correlation (R2 = 0.83) between the feature crosses and genomic characteristics was shown. The structural equation modeling confirmed significant associations between CT features, pathological (β = − 0.75), and molecular subtype (β = − 0.30).
Conclusions
The framework illustrates high performance in the pathological classification of renal cell carcinoma. Prognosis and genomic characteristics can be inferred by quantitative image analysis.
Key Points
• The analytical framework exhibits high-performance pathological classification of renal cell carcinoma and is on a par with human radiologists.
• Quantitative decomposition of the predictive model shows that specific texture features contribute to histologic subtype and tumor stage classification.
• Structural equation modeling shows the associations of genomic characteristics to CT texture features. Overall survival and molecular characteristics can be inferred by quantitative CT texture analysis in renal cell carcinoma.




Similar content being viewed by others
Data availability
All of the code generated or used during the study are available in the Github repository (https://github.com/wukaiyeah/kidney_cancer_project). The original images and data used in this study are available from the corresponding author by request.
Abbreviations
- p :
-
p Value
- SEM:
-
Structural equation modeling
- SHAP:
-
SHapley Additive exPlanations
- vs.:
-
Versus
- β:
-
Standardized beta coefficient
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69:7–34
Cancer Genome Atlas Research Network (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499:43–49
Feng X, Zhang L, Tu W, Cang S (2019) Frequency, incidence and survival outcomes of clear cell renal cell carcinoma in the United States from 1973 to 2014: A SEER-based analysis. Medicine (Baltimore) 98:1–9
Delahunt B, Eble JN, Egevad L, Samaratunga H (2019) Grading of renal cell carcinoma. Histopathology 74:4–17
Dechet CB, Zincke H, Sebo TJ et al (2003) Prospective analysis of computerized tomography and needle biopsy with permanent sectioning to determine the nature of solid renal masses in adults. J Urol 169:71–74
Silverman SG, Gan YU, Mortele KJ, Tuncali K, Cibas ES (2006) Renal masses in the adult patient: the role of percutaneous biopsy. Radiology 240:6–22
Ardila D, Kiraly AP, Bharadwaj S et al (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25:954–961
Nagpal K, Foote D, Liu Y et al (2019) Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2:1–10
McKinney SM, Sieniek M, Godbole V et al (2020) International evaluation of an AI system for breast cancer screening. Nature 577:89–94
Zhou L, Zhang Z, Chen Y-C, Zhao Z-Y, Yin X-D, Jiang H-B (2019) A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Trans Oncol 12:292–300
Deng Y, Soule E, Samuel A et al (2019) CT texture analysis in the differentiation of major renal cell carcinoma subtypes and correlation with Fuhrman grade. Eur Radiol 29:6922–6929
Kocak B, Yardimci AH, Bektas CT et al (2018) Textural differences between renal cell carcinoma subtypes: machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur J Radiol 107:149–157
Ding J, Xing Z, Jiang Z et al (2018) CT-based radiomic model predicts high grade of clear cell renal cell carcinoma. Eur J Radiol 103:51–56
Yang G, Gong A, Nie P et al (2019) Contrast-enhanced CT texture analysis for distinguishing fat-poor renal angiomyolipoma from chromophobe renal cell carcinoma. Mol Imaging 18:1536012119883161
Yu H, Scalera J, Khalid M et al (2017) Texture analysis as a radiomic marker for differentiating renal tumors. Abdom Radiol (NY) 42:2470–2478
Cui EM, Lin F, Li Q et al (2019) Differentiation of renal angiomyolipoma without visible fat from renal cell carcinoma by machine learning based on whole-tumor computed tomography texture features. Acta Radiol 60:1543–1552
Zhou M, Leung A, Echegaray S et al (2018) Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology 286:307–315
Iwatate Y, Hoshino I, Yokota H et al (2020) Radiogenomics for predicting p53 status, PD-L1 expression, and prognosis with machine learning in pancreatic cancer. Br J Cancer 123:1253–1261
Kocak B, Durmaz ES, Ates E, Ulusan MB (2019) Radiogenomics in clear cell renal cell carcinoma: machine learning–based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. AJR Am J Roentgenol 212:W55–W63
Yeh AC, Li H, Zhu Y et al (2019) Radiogenomics of breast cancer using dynamic contrast enhanced MRI and gene expression profiling. Cancer Imaging 19:48
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting systemproceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. Association for Computing Machinery, San Francisco, California, USA, pp 785–794
Lemaître G, Nogueira F, Aridas CK (2017) Imbalanced-learn: a python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mach Learn Res 18:559–563
Gibbons DL, Creighton CJ (2018) Pan-cancer survey of epithelial–mesenchymal transition markers across the Cancer Genome Atlas. Dev Dyn 247:555–564
Lundberg SM, Erion G, Chen H et al (2020) From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2:56–67
Motzer RJ, Jonasch E, Michaelson MD et al (2019) NCCN guidelines insights: kidney cancer, version 2.2020: featured updates to the NCCN guidelines. J Natl Compr Canc Netw 17:1278–1285
Kim BJ, Kim JH, Kim HS, Zang DY (2017) Prognostic and predictive value of VHL gene alteration in renal cell carcinoma: a meta-analysis and review. Oncotarget 8:13979
Zhao Y, Chang M, Wang R et al (2020) Deep learning based on MRI for differentiation of low-and high-grade in low-stage renal cell carcinoma. J Magn Reson Imaging 52:1542–1549
Trebeschi S, van Griethuysen JJM, Lambregts DMJ et al (2017) Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep 7:1–9
Coudray N, Ocampo PS, Sakellaropoulos T et al (2018) Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567
Kocak B, Kaya OK, Erdim C, Kus EA, Kilickesmez O (2020) Artificial intelligence in renal mass characterization: a systematic review of methodologic items related to modeling, performance evaluation, clinical utility, and transparency. AJR Am J Roentgenol 215:1113–1122
Maier-Hein L, Eisenmann M, Reinke A et al (2018) Why rankings of biomedical image analysis competitions should be interpreted with care. Nat Commun 9:1–13
Kocak B, Durmaz ES, Erdim C, Ates E, Kaya OK, Kilickesmez O (2020) Radiomics of renal masses: systematic review of reproducibility and validation strategies. AJR Am J Roentgenol 214:129–136
Yang W, Huang H, Zhang Z, Chen X, Huang K, Zhang S (2019) Towards rich feature discovery with class activation maps augmentation for person re-identification. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)
Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ (2017) CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 37:1483–1503
Taguchi N, Oda S, Yokota Y et al (2019) CT texture analysis for the prediction of KRAS mutation status in colorectal cancer via a machine learning approach. Eur J Radiol 118:38–43
Xu F, Ma X, Wang Y et al (2018) CT texture analysis can be a potential tool to differentiate gastrointestinal stromal tumors without KIT exon 11 mutation. Eur J Radiol 107:90–97
Sun R, Limkin EJ, Vakalopoulou M et al (2018) A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 19:1180–1191
Suarez-Ibarrola R, Basulto-Martinez M, Heinze A, Gratzke C, Miernik A (2020) Radiomics applications in renal tumor assessment: a comprehensive review of the literature. Cancers 12:1387
Acknowledgements
We thank the open-source database such as The Cancer Imaging Archive (TCIA)/The Cancer Genome Atlas (TCGA) for their data to this project.
Funding
This study has received funding from the National Natural Science Foundation of China (61931024 and 81922046), the National Key Research and Development Program of China (2017YFA0105900), the Special Funds for Strategic Emerging Industries Development in Shenzhen (20180309163446298), Shenzhen Key Laboratory Program (ZDSYS20190902092857146), and the Open Research Fund from Shenzhen Research Institute of Big Data (2019ORF01007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Pro. Song Wu, who is the Head of the Institute of Urology, Shenzhen University, China. Email address: wusong@szu.edu.cn
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
Kai Wu and Peng Wu, two of the authors, majored in statistics and computer sciences, have significant statistical expertise.
Informed consent
Informed consent documents are waived for all the participants are from an open-source project.
Ethical approval
This study was approved by the institutional research ethics committee. All data used were acquired with institutional review board-approved protocols.
Study subjects or cohorts overlap
All the cohorts have been previously reported in The Cancer Imaging Archive (TCIA), which is an open-source database and hosts a large archive of medical images accessible for public download. As far as we know, seldom researches integrated and analyzed all the kidney-cancer cohorts provided by TCIA. In this study, we proposed an analytical procedure by using these CT images, and firstly reported the correlation and causality between CT texture features and genomics in kidney cancer.
Methodology
• retrospective
• diagnostic or prognostic study / observational
• multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kai Wu and Peng Wu contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, K., Wu, P., Yang, K. et al. A comprehensive texture feature analysis framework of renal cell carcinoma: pathological, prognostic, and genomic evaluation based on CT images. Eur Radiol 32, 2255–2265 (2022). https://doi.org/10.1007/s00330-021-08353-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08353-3