Abstract
Objectives
To train a deep learning model to differentiate between pathologically proven hepatocellular carcinoma (HCC) and non-HCC lesions including lesions with atypical imaging features on MRI.
Methods
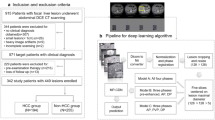
This IRB-approved retrospective study included 118 patients with 150 lesions (93 (62%) HCC and 57 (38%) non-HCC) pathologically confirmed through biopsies (n = 72), resections (n = 29), liver transplants (n = 46), and autopsies (n = 3). Forty-seven percent of HCC lesions showed atypical imaging features (not meeting Liver Imaging Reporting and Data System [LI-RADS] criteria for definitive HCC/LR5). A 3D convolutional neural network (CNN) was trained on 140 lesions and tested for its ability to classify the 10 remaining lesions (5 HCC/5 non-HCC). Performance of the model was averaged over 150 runs with random sub-sampling to provide class-balanced test sets. A lesion grading system was developed to demonstrate the similarity between atypical HCC and non-HCC lesions prone to misclassification by the CNN.
Results
The CNN demonstrated an overall accuracy of 87.3%. Sensitivities/specificities for HCC and non-HCC lesions were 92.7%/82.0% and 82.0%/92.7%, respectively. The area under the receiver operating curve was 0.912. CNN’s performance was correlated with the lesion grading system, becoming less accurate the more atypical imaging features the lesions showed.
Conclusion
This study provides proof-of-concept for CNN-based classification of both typical- and atypical-appearing HCC lesions on multi-phasic MRI, utilizing pathologically confirmed lesions as “ground truth.”
Key Points
• A CNN trained on atypical appearing pathologically proven HCC lesions not meeting LI-RADS criteria for definitive HCC (LR5) can correctly differentiate HCC lesions from other liver malignancies, potentially expanding the role of image-based diagnosis in primary liver cancer with atypical features.
• The trained CNN demonstrated an overall accuracy of 87.3% and a computational time of < 3 ms which paves the way for clinical application as a decision support instrument.





Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- CNN:
-
Convolutional neural network
- FNH:
-
Focal nodular hyperplasia
- HCC:
-
Hepatocellular carcinoma
- HIPAA:
-
Health Insurance Portability and Accountability Act
- ICC:
-
Intrahepatic cholangiocarcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- MELD:
-
Model for End-Stage Liver Disease
- NPV:
-
Negative predictive value
- PACS:
-
Picture archiving and communication system
- PPV:
-
Positive predictive value
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Wald C, Russo MW, Heimbach JK, Hussain HK, Pomfret EA, Bruix J (2013) New OPTN/UNOS policy for liver transplant allocation: standardization of liver imaging, diagnosis, classification, and reporting of hepatocellular carcinoma. Radiology 266:376–382. https://doi.org/10.1148/radiol.12121698
CT/MRI LI-RADS v2018. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2018. Accessed 31 Aug 2020
Davenport MS, Khalatbari S, Liu PSC et al (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272:132–142. https://doi.org/10.1148/radiol.14131963
Smith EH (1991) Complications of percutaneous abdominal fine-needle biopsy. Review. Radiology 178:253–258. https://doi.org/10.1148/radiology.178.1.1984314
Seehofer D, Öllinger R, Denecke T et al (2017) Blood transfusions and tumor biopsy may increase HCC recurrence rates after liver transplantation. J Transplant. https://doi.org/10.1155/2017/9731095
Quaia E, De Paoli L, Angileri R, Cabibbo B, Cova MA (2014) Indeterminate solid hepatic lesions identified on non-diagnostic contrast-enhanced computed tomography: assessment of the additional diagnostic value of contrast-enhanced ultrasound in the non-cirrhotic liver. Eur J Radiol 83:456–462. https://doi.org/10.1016/j.ejrad.2013.12.012
Pérez Saborido B, Menéu Díaz JC, Jiménez de los Galanes S et al (2005) Does preoperative fine needle aspiration-biopsy produce tumor recurrence in patients following liver transplantation for hepatocellular carcinoma? Transplant Proc 37:3874–3877. https://doi.org/10.1016/j.transproceed.2005.09.169
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging. https://doi.org/10.1007/s13244-018-0639-9
Greenspan H, van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159. https://doi.org/10.1109/TMI.2016.2553401
Hamm CA, Wang CJ, Savic LJ et al (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. https://doi.org/10.1007/s00330-019-06205-9
Wang CJ, Hamm CA, Savic LJ et al (2019) Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol 29:3348–3357. https://doi.org/10.1007/s00330-019-06214-8
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896. https://doi.org/10.1148/radiol.2017170706
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90. https://doi.org/10.1145/3065386
Erickson BJ, Korfiatis P, Akkus Z, Kline TL (2017) Machine learning for medical imaging. Radiographics 37:505–515. https://doi.org/10.1148/rg.2017160130
Arlot S, Celisse A (2010) A survey of cross-validation procedures for model selection. Statist Surv 4:40–79. https://doi.org/10.1214/09-SS054
CT/MRI LI-RADS v2017. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2017. Accessed 17 May 2018
He K, Zhang X, Ren S, Sun J (2016) Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, Maaten LVD, Weinberger KQ (2017) Densely Connected Convolutional Networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2261–2269. https://doi.org/10.1109/CVPR.2017.243
Fodor M, Primavesi F, Braunwarth E et al (2018) Indications for liver surgery in benign tumours. Eur Surg 50:125–131. https://doi.org/10.1007/s10353-018-0536-y
Funding
CW received funding from the Radiological Society of North America (RSNA Research Resident Grant #RR1731). JD, JC, ML, and CW received funding from the National Institutes of Health (NIH/NCI R01 CA206180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Julius Chapiro, MD, PhD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Oestmann, P.M., Wang, C.J., Savic, L.J. et al. Deep learning–assisted differentiation of pathologically proven atypical and typical hepatocellular carcinoma (HCC) versus non-HCC on contrast-enhanced MRI of the liver. Eur Radiol 31, 4981–4990 (2021). https://doi.org/10.1007/s00330-020-07559-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07559-1