Abstract
Objective
To assess significant liver fibrosis by multiparametric ultrasomics data using machine learning.
Materials and Methods
This prospective study consisted of 144 patients with chronic hepatitis B. Ultrasomics—high-throughput quantitative data from ultrasound imaging of liver fibrosis—were generated using conventional radiomics, original radiofrequency (ORF) and contrast-enhanced micro-flow (CEMF) features. Three categories of features were explored using pairwise correlation and hierarchical clustering. Features were selected using diagnostic tests for fibrosis, activity and steatosis stage, with the histopathological results as the reference. The fibrosis staging performance of ultrasomics models with combinations of the selected features was evaluated with machine-learning algorithms by calculating the area under the receiver-operator characteristic curve (AUC).
Results
ORF and CEMF features had better predictive power than conventional radiomics for liver fibrosis stage (both p < 0.01). CEMF features exhibited the highest diagnostic value for activity stage (both p < 0.05), and ORF had the best diagnostic value for steatosis stage (both p < 0.01). The machine-learning classifiers of adaptive boosting, random forest and support vector machine were found to be optimal algorithms with better (all mean AUCs = 0.85) and more stable performance (coefficient of variation = 0.01–0.02) for fibrosis staging than decision tree, logistic regression and neural network (mean AUC = 0.61–0.72, CV = 0.07–0.08). The multiparametric ultrasomics model achieved much better performance (mean AUC values of 0.78–0.85) than the features from a single modality in discriminating significant fibrosis (≥ F2).
Conclusion
Machine-learning-based analysis of multiparametric ultrasomics can help improve the discrimination of significant fibrosis compared with mono or dual modalities.
Key Points
• Multiparametric ultrasomics has achieved much better performance in the discrimination of significant fibrosis (≥ F2) than the single modality of conventional radiomics, original radiofrequency and contrast-enhanced micro-flow.
• Adaptive boosting, random forest and support vector machine are the optimal algorithms for machine learning.
Similar content being viewed by others
Introduction
The early detection and accurate staging of liver significant fibrosis are crucial for antiviral therapy. Shear wave elastography (SWE), an elasticity-based US technique, has shown good accuracy in detecting fibrosis [1]. However, the applicability of SWE is substantially limited in cases of obesity, ascites or necroinflammatory activity (up to 15.8%) [2, 3]. Thus, assessment with a single imaging modality only provides limited information and could always be affected by steatosis and necroinflammatory activity [4, 5].
Radiomics, a term that includes the suffix “-omics,” generates high-throughput data from medical images [6, 7], which contain information on prognosis, response to treatment and monitoring of disease status [8, 9]. As one important modality of medical imaging, US can provide not only morphological information but also stiffness and perfusion assessments, which may not be acquired using other imaging methods [5, 10,11,12]. We have applied the “-omics” concept to computing quantitative US imaging, a field referred to as “ultrasomics.” In our opinion, big imaging data of liver fibrosis, in terms of heterogeneity, tissue texture, stiffness and vascularity perfusion, should be taken into consideration when analyzing fibrosis staging.
In addition to multimodality data, machine learning is another powerful tool to improve clinical decision-making [13]. Currently, newer advances in data analysis contributed by the field of machine learning have greatly extended researchers’ ability to make meaningful discoveries. Machine learning enables accurate and reliable prediction using data with very large numbers of variables and small sample sizes. Therefore, the optimal machine-learning model for ultrasomics studies with small sample sizes should be determined. To our knowledge, comparative studies on the effectiveness of machine learning-based decision support systems are lacking [14].
In this study, we present the concept of multiparametric ultrasomics, which is a machine learning-based clinical decision support system that uses US imaging big data. We extracted a set of ultrasomic features that captures the morphology and hemodynamic changes associated with liver fibrosis to (1) develop a robust, noninvasive technique to predict the liver fibrosis stage using routine US data that can be easily obtained in the clinical setting and (2) investigate the optimal machine-learning model in a small sample size study.
Materials and methods
Study population
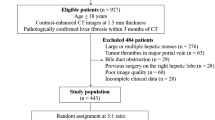
This prospective study was approved by the Institutional Review Board of our hospital, and informed consent was obtained. From October 2013 to April 2015, a total of 144 hepatitis B virus (HBV) patients who met the eligibility criteria were included in the study. The inclusion and exclusion criteria are detailed in the Supplementary Materials. For each patient, comprehensive blood tests (aspartate transaminase (AST), alanine transaminase (ALT), serum albumin, g-glutamyltransferase, total bilirubin, and platelet count) were performed no more than 3 days before the surgery or biopsy. Combinations of simple markers such as the aspartate aminotransferase-to-platelet ratio index (APRI) and fibrosis-4 index (FIB-4) were calculated; the formula is provided in the Supplementary Materials [15].
Liver histology analysis
All patients included in the study underwent partial liver surgery (n = 61) or biopsy (n = 83). Resected liver specimens approximately 10 mm × 10 mm in size were preserved intraoperatively. A US-guided percutaneous liver biopsy of the right lobe was performed with an 18-gauge needle (Bard) within 3 days after ultrasonography. All specimens were fixed in formalin, embedded in paraffin and stained with hematoxylin-eosin (H&E) and Masson. Two liver pathologists with > 10 years of experience, who were blinded to the results of imaging but not to the clinical and biochemical data of the patient, analyzed the specimens. Liver fibrosis was evaluated according to the METAVIR scoring system as follows: F0, no fibrosis; F1, portal fibrosis without septa; F2, portal fibrosis and few septa; F3, numerous septa without cirrhosis; F4, cirrhosis. Significant fibrosis was defined as a score of F2 or greater. The METAVIR system was used to score the intensity of necroinflammatory activity (mainly necrosis) as follows: A0 = no necroinflammatory activity, A1 = mild activity, A2 = moderate activity and A3 = severe activity [4]. Steatosis was scored, using a four-grade scoring system, from S0 to S4: S0 = no steatosis; S1 = mild (1–5%) (% of hepatocytes containing visible macrovesicular steatosis); S2 = moderate (6–32%); S3 = marked (33–66%); S4 = severe (67–100%) [4, 16, 17].
Multiparametric ultrasomics acquisition and feature extraction
All US examinations were performed with the Aplio 500 scanner (Canon Medical System) equipped with a 375BT convex transducer (frequency, 3.5 MHz). US examinations were performed by one of two radiologists (X.Y.X. and W.W.) with at least 10 years of experience with routine US. Three types of parameters were acquired:
-
1.
B-mode images in digital imaging and communications in medicine (DICOM) format. Images were obtained with intercostal oblique scanning and were expected to show the liver parenchyma from the right intercostal space to the segment 6 region of the right hepatic lobe. Display depth and transmit focus were fixed at 6 cm and 4 cm, respectively, with the receive gain equal to 80. Large vessels defined as > 2.0 mm were avoided. The settings, including time-gain compensation, dynamic range, focal length and mechanical index, were optimized for each examination. Conventional images in DICOM format were stored on the Canon Medical System platform. DICOM images were used to extract conventional radiomics features using A. K. software (Ultrasomics Kit, version 1.0, ZhiXing-Tech), including the first-order intensity statistics, texture and wavelet features. Mathematical definitions of all radiomic features were previously described and are detailed in the Supplementary Materials [18].
-
2.
Radiofrequency-based raw data. The same scanning method and planes of conventional radiomics were used, but data were stored as raw data. Post-beam-formed original radiofrequency data (ORF features) with intact frequency information were used to extract the statistical features of the acquired echo amplitude of the raw data.
-
3.
Dynamic contrast-enhanced micro-flow (CEMF) images. Contrast harmonic imaging was used with a mechanical index of 0.08. The transmission frequency used in contrast harmonic imaging was 3.5 MHz and a frame rate of 15–18 frames per second. Images were obtained with intercostal oblique scanning and were expected to show the liver parenchyma from the right intercostal space to the segment 6 region of the right hepatic lobe and the right kidney on a single screen. Focus was set at a depth of 6 to 8 cm to visualize the kidney. After the contrast harmonic imaging mode was activated, a bolus injection of 2.4 ml of SonoVue (Bracco) was administered intravenously via an antecubital vein, followed immediately by a 5 ml saline flush. Patients were instructed to hold their breath after the injection for 7–8 s (1–2 s before the visualization of renal artery), and then the CEMF mode was initiated. Image acquisition proceeded until the liver was wholly enhanced. Clips obtained for approximately 15–20 s immediately after SonoVue infusion were saved as clip data in DICOM format. Dynamic CEMF features were extracted via our built-in model through off-line analysis. The model was developed with the understanding that the liver received dual blood supply from the hepatic artery and portal vein. The blood supply of the kidney was set as the “hepatic artery supply” and was used as a comparison indicator to reduce the influence of circulation difference.
According to our preliminary and reported studies [10, 11, 19,20,21], the three categories of parameters (conventional radiomics, ORF and CEMF features) acquired from the three types of images were expected to provide potential information for liver fibrosis staging and are detailed in the Supplementary Materials. In total, there were 472 features, including 396 conventional radiomics, 54 ORF and 22 CEMF features. These computed quantitative features were selected to construct the ultrasomics—the “omics” data of ultrasound in this study (Fig. 1).
The diagram shows the four-step process for the construction of an ultrasomic-based predictive model. (I) Ultrasomic images were obtained from different US modalities (conventional images, original radiofrequency data and dynamic CEMF images). (II) Big ultrasonic data were extracted as conventional radiomic features, original radiofrequency features and dynamic CEMF features. (III) Big data mining (correlation, cluster and predictive performance) was performed to select the optimal predictor, and the classification performance of ultrasomic models was tested via machine learning. (IV) In this study, we prospectively enrolled 144 patients with liver fibrosis to establish an ultrasomic model for the prediction of fibrosis stages
Multiparameter-based ultrasomic analysis of liver fibrosis using machine learning
Feature selection and analysis of multiparametric ultrasomics
Spearman’s correlation coefficient (R) was used to assess correlations between features in all parameters. Feature pairs with |R| > 0.90 were considered to be highly correlated and likely to provide redundant rather than complementary information. The highly correlated features were collapsed into one representative feature, usually the one with the greatest variability or highest dynamic range. This procedure yielded independent features for conventional radiomics, ORF and CEMF features.
Then, we explored feature correlations by establishing a correlation map for pairwise associations among the three categories of parameters. A hierarchical cluster of all quantitative features was plotted with different stages of fibrosis, activity and steatosis. The performance of each feature was further quantified by calculating the area under the receiver-operator characteristic curve (AUC) for fibrosis stage, activity and steatosis.
Multiparametric ultrasomic-based models for significant fibrosis using machine learning
Three categories of parameters with AUCs > 0.6 for assessing all stages of fibrosis were selected for the following analysis. To assess the optimal machine-learning method, all parameters of the three categories were selected for model construction. A total of six types of machine-learning algorithms—adaptive boosting (AdaBoost), decision tree (DT), logistic regression (LR), neural network (NN), random forest (RF) and support vector machine (SVM)—were tested in this study. These machine-learning algorithms were selected because of their promising performance in classification [13]. The brief descriptions of each classifier were explained in the Supplementary Material.
The entire cohort was randomly divided into a training data set (100 cases) and validation data set (44 cases). The training data set was used to compose a model and evaluated it by a validation data set. Six models with different machine-learning methods were built on the training data set, and the performance of each model was then assessed on the validation data set. To ensure the robustness of the classifiers to training and testing data, we adopted a ten-fold cross-validation method to calculate the diagnostic value for significant fibrosis (≥ F2). All processes were repeated ten times with random seed, resulting in ten different training and validation data sets. We repeatedly composed a model using a training data set and evaluated it by a validation data set, and a model that showed the best classification performance was chosen as the best model. The classification performance for significant fibrosis was assessed using the AUC in the validation data sets.
Comparison of multiparametric ultrasomics models
Multiparametric ultrasomics models using optimized machining-learning methods were compared against models of three categories of parameters combined: (1) a combination of conventional radiomics, ORF and CEMF; two categories of parameters combined: (2) a combination of conventional radiomics and CEMF, (3) a combination of ORF and CEMF, (4) a combination of conventional radiomics and ORF; and models with a single parameter: (5) conventional radiomics, (6) ORF and (7) CEMF. Classifier performance was assessed by computing the accuracy, sensitivity, specificity and receiver-operating characteristic (ROC) curve. The AUCs for significant fibrosis in the validation data sets were assessed with the adopted ten-fold cross-validation method.
Statistical analysis
Statistical analyses were performed with the open-source statistical computing environment R (version 3.3.1; R Foundation for Statistical Computing). We filtered features of three modalities based on independence from other features (intraclass Pearson correlation, |r| > 0.9). Heat maps of interclass Pearson correlations among the three categories of parameters were calculated and plotted using the R package “corrplot”. Six machine-learning algorithms were applied with the R packages “rpart”, “ada”, “randomForest”, “kernlab”, “rms” and “nnet”. AUCs for staging fibrosis, activity and steatosis were explored for the three categories of features with the R package “pROC”. The differences between model performance across different machine-learning and parameter subgroups were evaluated via a permutation test by using the R package “Deducer”. Coefficients of variation (CVs) were calculated to compare the discrete degrees of AUCs. All statistical tests were two-sided, and p values < 0.05 were considered statistically significant.
Results
Patient characteristics
A total of 144 patients were enrolled in the study, of whom 114 were male. The mean BMI was 20 kg/m2, and the mean age was 48 years. METAVIR fibrosis, activity and steatosis distribution are summarized in Table 1.
Feature selection and analysis of multiparametric ultrasomics
The independent features for conventional radiomics, ORF and CEMF were 110, 20 and 26, respectively. In general, these features have barely no correlation between ORF and CEMF (Spearman rho, -0.26 to 0.24) and between CEMF and conventional radiomics (Spearman rho, -0.26 to 0.32); however, the correlation coefficient of ORF and conventional radiomics features was slightly correlated and higher than in the above two groups (Spearman rho, -0.34 to 0.47) (Fig. 2).
The correlation heat map shows associations between conventional radiomics, ORF and CEMF features. Only a few parameters were highly correlated (blue) or highly anti-correlated (red). In general, we found that these features were only slightly or moderately correlated between ORF and CEMF features (a, Spearman rho, -0.26 to 0.24) and CEMF and conventional radiomic features (b, Spearman rho, -0.26 to 0.32). However, the variables between ORF and conventional radiomic features were more highly correlated (c, Spearman rho, -0.34 to 0.47)
Hierarchical clusters of all quantitative features were plotted with different stages of fibrosis, activity and steatosis (Fig. S1). We tested the diagnostic value of the selected parameters for fibrosis, activity and steatosis stage using AUC. A boxplot showed that ORF and CEMF features were the strongest predictors, with no significant difference, and were much stronger than conventional radiomics features for all fibrosis stages (Fig. 3a, both p < 0.01). Additionally, CEMF features exhibited the highest diagnostic value for all activity stages (Fig. 3b), and ORF performed the best for all steatosis stages (Fig. 3c).
Diagnostic value of the parameters in the diagnosis of fibrosis, activity and steatosis stages. The boxplot shows that ORF and CEMF parameters, with no significant difference, were the strongest predictors and were much higher than conventional radiomic features for liver fibrosis stages (a, both p < 0.01, ANOVA test). CEMF exhibited the highest diagnostic value for activity stages (b, both p < 0.05, ANOVA test), and ORF performed the best for steatosis stages (c, both p < 0.01, ANOVA test)
Multiparametric ultrasomic-based models for significant fibrosis using machine learning
Of the 156 features analyzed, 93 variables for conventional radiomics features, 11 variables for ORF features and 11 variables for CEMF features were removed in the final models because of their low diagnostic value (AUC < 0.6). The model for machine learning included 41 features: 17 conventional radiomics features, 15 ORF and 9 CEMF features. The detailed name and definitions of these features were listed in the Supplementary Material.
The classification performance of each of the six machine-learning classification methods is shown in Fig. 4. The results showed that the AdaBoost, RF and SVM classifiers (mean AUC = 0.85 for all three) outperformed the other classifiers (mean AUC = 0.61–0.72, all p < 0.001). In addition, these three classifiers showed less variation, with a CV of 0.01–0.02, which was much lower than those of the other models (CV = 0.07–0.08). Based on these findings, the AdaBoost, RF and SVM classifiers were selected for the next computing because of their stability, effectiveness and high staging performance.
Boxplot showing the classification performance with the six machine-learning methods with all parameters. The p value is for a permutation test. The AdaBoost, RF and SVM classifier outperformed the other classifiers (all p < 0.001). These three classifiers showed less variation with a smaller quartile value and dispersion degree
Comparison of multiparametric ultrasomics models
The performance of machine-learning prediction models for each parameter group is summarized in Fig. 5 and Tables 2 and 3. As seen from this distribution, the combination of multiparametric features achieved much better performance (mean AUC of 0.78–0.85) than the features from a single modality (mean AUC of 0.68–0.77). For models incorporating multiparametric conventional radiomics, ORF and CEMF, classifiers of AdaBoost, RF and SVM demonstrated good performance, with the same AUC of 0.85 ± 0.01 (with 87.5%, 87.5% and 93.8% sensitivity and 76.9%, 76.9% and 69.2% specificity, respectively). For models combining two modalities, combinations of ORF and CEMF and combinations of conventional radiomics and CEMF demonstrated good performance, with mean AUCs of 0.82–0.85; however, the performance of the models combining conventional radiomics and ORF was poorer, with a mean AUC of 0.78–0.79. The models with a single modality showed only fair performance, with a mean AUC of 0.68–0.77.
The results of three machine-learning method analyses in classifying each combination of ultrasomic features. All three machine-learning methods-adaptive boosting (a), random forest (b) and support vector machine (c) showed that the ultrasomics models achieved much better performance than the models of a single modality and models of conventional radiomics and ORF features (all p < 0.001)
Discussion
In the current study, we propose the use of multiparametric ultrasomics as a decision support tool for liver fibrosis staging. In addition to conventional radiomics features from digital images, we acquired RF signal and dynamic perfusion information to construct ultrasomics, which are unique but convenient to acquired ultrasound parameters [10, 11, 20]. These mineable data for the evaluation of fibrosis staging were tested and compared with different machine-learning algorithms. Multiparametric ultrasomics using AdaBoost, RF and SVM provided the highest performance in this study with a small sample size.
In the construction of ultrasomics, we used unsupervised machine learning to explore the data characteristics of the parameters. A higher correlation between ORF and conventional radiomics features was found. An ORF signal is post-beam-formed data from a transducer, and it can provide intact information without signal processing [22]. Radiomics parameters are conventional features that are mathematically extracted quantitative descriptors based on digitally transformed images [23]. Although the digital images were signals post-processed with a digital scan converter, both signals simulated the morphological homogeneity of liver tissue. Notably, the ORF data, which included original and superior information, performed better in the assessment of fibrosis and steatosis. Some papers have reported results on the tissue characterization of hepatic fibrosis or steatosis via quantitative ultrasound examination using statistical data on B-mode ultrasound and radiofrequency echo signals [24,25,26]. However, dynamic perfusion parameters, showing a lower correlation with both morphological parameters, demonstrated the highest diagnostic value for activity stages, which was correlated with liver microcirculation [27]. Therefore, our results suggested that these three signals could be divided into two categories: morphology and hemodynamics, which reflected the fibrosis and steatosis stages and the activity stages, respectively.
For liver fibrosis staging with ultrasomics, although the optimized machine-learning algorithms had been selected, a clinical model that used single-modality parameters still provided unsatisfactory AUC values for staging (AUC < 0.8). In addition, the models that used duplicate morphological parameters (higher correlated features of ORF and conventional radiomics) displayed the lowest AUCs in the validation groups, which may be due to the redundant information between two morphological parameters. Moreover, the models using combined morphology and hemodynamic features demonstrated better performance. For the evaluation of fibrosis stage, the accompanying activity of liver tissue should not be ignored [28, 29]. Our results also agree with the principle that models constructed with two categories should achieve higher AUCs despite the use of a machine-learning algorithm. This finding shows that the use of multiparametric ultrasomics from different pathophysiological procedures would enhance the performance of our clinical decision support system [30].
With these big data, machine learning is driving great changes in medical disciplines that are based on pattern recognition (e.g., radiology and pathology) [31, 32]. Generally, machine learning with a larger sample size produces more accurate classification [32,33,34]. However, the ultrasound images that qualified for the computing analysis in the present work were restricted because of their dependence on the operator and software, and our study included only 144 cases as a result. Therefore, determining the optimal machine-learning algorithm for a small sample size is of great value. A few recent studies have investigated the effects of different machine-learning classification methods on single-modality radiomic-based clinical predictions [35, 36]. Our study showed that the three machine-learning methods of AdaBoost, RF and SVM performed better with any category of parameters. The principle of SVM is to map the input parameters into a high-dimensional feature space via preselected nonlinear mapping [33, 37]. In this space, an optimal classification hyperplane is constructed and is optimized to maximize the classification of the two categories. The use of a margin among the hyperplane and two categories reduces the size and distribution requirements of the data. RF combines predictions from several weak classifiers to generate a more accurate and stable prediction. Random samples and features guarantee the robustness to noise in the data with few tuning parameters and a small sample size [32, 38, 39]. AdaBoost is adaptive in the sense that subsequent weak learners are tweaked in favor of instances misclassified by previous classifiers [13, 36]. Thus, AdaBoost is sensitive to noisy data and outliers; this sensitivity makes AdaBoost less susceptible to the overfitting problem.
The first and most important limitation of this study was that multiparametric ultrasomics could not include parameters of liver stiffness. Many single-center studies have already shown that shear wave elastography achieved a good AUC of 0.89 in staging F2 fibrosis patients [17]. However, considering patient compliance, only one US machine was used to acquire the ultrasound data in this prospective study. Additionally, the primary purpose of this study was to optimize the model of fibrosis staging based on multiparametric ultrasomics using machine learning. Our present work has demonstrated that (1) data exploration correlating with pathophysiology and (2) model construction using machine learning could improve the robustness of fibrosis staging models. However, the reported performance of shear wave elastography in staging significant fibrosis (AUC = 0.69–0.92, sensitivity = 0.77–0.90 and specificity = 0.70–0.87) was similar to that of our multiparametric model. This finding reminds us that, although the data mining process could enhance the performance of clinical decision systems, the optimal performance of the model has already been determined by the data. In a subsequent study, we will collect shear wave elastography data to produce a better model.
Our study also had other limitations. Second, our study only included a cohort of patients in our hospital. It is necessary to establish an independent validation cohort to test the generalizability of our ultrasomics model. A third limitation is that several patients enrolled in the study had focal liver lesions. This may also affect the association between US parameters and pathology. To eliminate the potential effect of tumors on the adjacent liver parenchyma, we created strict exclusion criteria. Fourth, the population of the study was Chinese patients with chronic HBV, which led to a low BMI of 20.2 kg/m2 and a low proportion of liver steatosis. Fifth, due to the requirement in image acquisition, only a short time (15–20-s clips) was covered in the CEMF images. This may not show the whole perfusion procedure in the liver. However, we attempted to analyze the blood flow arrival time to the liver and kidney based on a time-intensity curve, and the wash-in curve contains more information for the arrival time. Sixth, the analysis of the liver (conventional radiomics and ORF) was done from a small 2-cm ROI placed in segment 6, which has the same limitation as a biopsy of not reflecting the potential heterogeneity of the liver disease.
In summary, we have demonstrated that expert knowledge on data acquisition and analysis can optimize the robustness of clinical decision support systems. Additionally, the three machine-learning methods of AdaBoost, RF and SVM are optimal algorithms for studies with a small sample size. The application of this framework in future studies will facilitate data mining in the era of ultrasomics.
Abbreviations
- AdaBoost:
-
Adaptive boosting
- CEMF:
-
Contrast enhanced micro-flow
- CV:
-
Coefficient of variation
- DT:
-
Decision tree
- LR:
-
Logistic regression
- NN:
-
Neural network
- ORF:
-
Original radiofrequency
- RF:
-
Random forest
- SVM:
-
Support vector machine
References
Ferraioli G, Tinelli C, Dal Bello B et al (2012) Accuracy of real-time shear wave elastography for assessing liver fibrosis in chronic hepatitis C: a pilot study. Hepatology 56:2125–2133
Castéra L, Foucher J, Bernard PH et al (2010) Pitfalls of liver stiffness measurement: a 5-year prospective study of 13,369 examinations. Hepatology 51:828–835
Berzigotti A, Castéra L (2013) Update on ultrasound imaging of liver fibrosis. J Hepatol 59:180–182
Bedossa P, Poynard T (1996) An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 24:289–293
Corpechot C, Carrat F, Poujol-Robert A et al (2012) Noninvasive elastography-based assessment of liver fibrosis progression and prognosis in primary biliary cirrhosis. Hepatology 56:198–208
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Verma V, Simone CB 2nd, Krishnan S, Lin SH, Yang J, Hahn SM (2017) The rise of radiomics and implications for oncologic management. J Natl Cancer Inst 109
Huang Y, Liang C, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Huang Y, Wang Z, Liao B et al (2016) Assessment of liver fibrosis in chronic hepatitis B using acoustic structure quantification: quantitative morphological ultrasound. Eur Radiol 26:2344–2351
Li W, Wang W, Liu GJ et al (2015) Differentiation of atypical hepatocellular carcinoma from focal nodular hyperplasia: diagnostic performance of contrast-enhanced US and microflow imaging. Radiology 275:870–879
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol. https://doi.org/10.1007/s00330-018-5499-7
Jordan MI, Mitchell TM (2015) Machine learning: trends, perspectives, and prospects. Science 349:255–260
Chen JH, Asch SM (2017) Machine learning and prediction in medicine—beyond the peak of inflated expectations. N Engl J Med 376:2507–2509
Hasson H, Merli M, Galli L et al (2013) Non-invasive fibrosis biomarkers—APRI and Forns—are associated with liver stiffness in HIV-monoinfected patients receiving antiretroviral drugs. Liver Int 33:1113–1120
Poynard T, Ratziu V, McHutchison J et al (2003) Effect of treatment with peginterferon or interferon alfa-2b and ribavirin on steatosis in patients infected with hepatitis C. Hepatology 38:75–85
Deffieux T, Gennisson JL, Bousquet L et al (2015) Investigating liver stiffness and viscosity for fibrosis, steatosis and activity staging using shear wave elastography. J Hepatol 62:317–324
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Huang Y, Liu GJ, Liao B et al (2015) Impact factors and the optimal parameter of acoustic structure quantification in the assessment of liver fibrosis. Ultrasound Med Biol 41:2360–2367
Wakui N, Takayama R, Kanekawa T et al (2012) Usefulness of arrival time parametric imaging in evaluating the degree of liver disease progression in chronic hepatitis C infection. J Ultrasound Med 31:373–382
Daginawala N, Li B, Buch K et al (2016) Using texture analyses of contrast enhanced CT to assess hepatic fibrosis. Eur J Radiol 85:511–517
Lin CY, Cao LH, Wang JW et al (2013) Ultrasonic spectrum analysis for in vivo characterization of tumor microstructural changes in the evaluation of tumor response to chemotherapy using diagnostic ultrasound. BMC Cancer 13:302
Limkin E, Sun R, Dercle L et al (2017) Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 28:1191–1206
Yamada H, Ebara M, Yamaguchi T et al (2006) A pilot approach for quantitative assessment of liver fibrosis using ultrasound: preliminary results in 79 cases. J Hepatol 44:68–75
Fujii Y, Taniguchi N, Wang Y et al (2004) Clinical application of a new method that segments the region of interest into multiple layers for RF amplitude histogram analysis in the cirrhotic liver. J Med Ultrason (2001) 31:91–98
Thijssen JM, Starke A, Weijers G et al (2008) Computer-aided B-mode ultrasound diagnosis of hepatic steatosis: a feasibility study. IEEE Trans Ultrason Ferroelectr Freq Control 55:1343–1354
Lettmann KA, Hardtke-Wolenski M (2014) The importance of liver microcirculation in promoting autoimmune hepatitis via maintaining an inflammatory cytokine milieu—a mathematical model study. J Theor Biol 348:33–46
Nieminen U, Arkkila P, Kärkkäinen P, Färkkilä M (2009) Effect of steatosis and inflammation on liver fibrosis in chronic hepatitis C. Liver Int 29:153–158
Morishima C, Shiffman M, Dienstag J et al (2012) Reduction in hepatic inflammation is associated with less fibrosis progression and fewer clinical outcomes in advanced hepatitis C. Am J Gastroenterol 107:1388–1398
Collij LE, Heeman F, Kuijer JP et al (2016) Application of machine learning to arterial spin labeling in mild cognitive impairment and Alzheimer disease. Radiology 281:865–875
Hodgdon T, McInnes MD, Schieda N, Flood TA, Lamb L, Thornhill RE (2015) Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology 276:787–796
Ueno Y, Forghani B, Forghani R et al (2017) Endometrial carcinoma: MR imaging-based texture model for preoperative risk stratification—a preliminary analysis. Radiology 284:748–757
Möller C, Pijnenburg YA, van der Flier WM et al (2016) Alzheimer disease and behavioral variant frontotemporal dementia: automatic classification based on cortical atrophy for single-subject diagnosis. Radiology 279:838–848
Burns J, Yao J, Summers R (2017) Vertebral Body compression fractures and bone density: automated detection and classification on CT images. Radiology 284:788–797
Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ (2015) Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol 5:272
Zhang B, He X, Ouyang F et al (2017) Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett 403:21–27
Bryan RN (2016) Machine learning applied to Alzheimer disease. Radiology 281:665–668
Agosta F, Ferraro PM, Canu E et al (2015) Differentiation between subtypes of primary progressive aphasia by using cortical thickness and diffusion-tensor MR imaging measures. Radiology 276:219–227
Preziosa P, Rocca MA, Mesaros S et al (2014) Relationship between damage to the cerebellar peduncles and clinical disability in multiple sclerosis. Radiology 271:822–830
Funding
This study was supported by the National Natural Science Foundation of China (nos. 81471672, 81701701 and 81701719) and the Guangdong Science and Technology Foundation (nos. 2016A030310143, 2016A020215042 and 2015A030310144).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Wei Wang.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• prospective
• diagnostic or prognostic study
• performed at one institution
Electronic supplementary material
ESM 1
(DOCX 3527 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, W., Huang, Y., Zhuang, BW. et al. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol 29, 1496–1506 (2019). https://doi.org/10.1007/s00330-018-5680-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5680-z