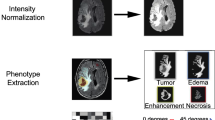
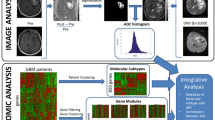
Abstract
Objective
To develop and validate a volume-based, quantitative imaging marker by integrating multi-parametric MR images for predicting glioblastoma survival, and to investigate its relationship and synergy with molecular characteristics.
Methods
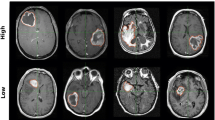
We retrospectively analysed 108 patients with primary glioblastoma. The discovery cohort consisted of 62 patients from the cancer genome atlas (TCGA). Another 46 patients comprising 30 from TCGA and 16 internally were used for independent validation. Based on integrated analyses of T1-weighted contrast-enhanced (T1-c) and diffusion-weighted MR images, we identified an intratumoral subregion with both high T1-c and low ADC, and accordingly defined a high-risk volume (HRV). We evaluated its prognostic value and biological significance with genomic data.
Results
On both discovery and validation cohorts, HRV predicted overall survival (OS) (concordance index: 0.642 and 0.653, P < 0.001 and P = 0.038, respectively). HRV stratified patients within the proneural molecular subtype (log-rank P = 0.040, hazard ratio = 2.787). We observed different OS among patients depending on their MGMT methylation status and HRV (log-rank P = 0.011). Patients with unmethylated MGMT and high HRV had significantly shorter survival (median survival: 9.3 vs. 18.4 months, log-rank P = 0.002).
Conclusion
Volume of the high-risk intratumoral subregion identified on multi-parametric MRI predicts glioblastoma survival, and may provide complementary value to genomic information.
Key points
• High-risk volume (HRV) defined on multi-parametric MRI predicted GBM survival.
• The proneural molecular subtype tended to harbour smaller HRV than other subtypes.
• Patients with unmethylated MGMT and high HRV had significantly shorter survival.
• HRV complements genomic information in predicting GBM survival





Similar content being viewed by others
References
Ostrom QT, Gittleman H, Fulop J et al (2015) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol 17(Suppl 4):iv1–iv62
Verhaak RG, Hoadley KA, Purdom E et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110
Jain R, Poisson L, Narang J et al (2013) Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 267:212–220
Burth S, Kickingereder P, Eidel O et al (2016) Clinical parameters outweigh diffusion- and perfusion-derived MRI parameters in predicting survival in newly diagnosed glioblastoma. Neuro Oncol. doi:10.1093/neuonc/now122
Schmainda KM, Zhang Z, Prah M et al (2015) Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro Oncol 17:1148–1156
Choi YS, Ahn SS, Kim DW et al (2016) Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology. doi:10.1148/radiol.2016151913:151913
Gupta A, Prager A, Young RJ, Shi W, Omuro AM, Graber JJ (2013) Diffusion-weighted MR imaging and MGMT methylation status in glioblastoma: a reappraisal of the role of preoperative quantitative ADC measurements. AJNR Am J Neuroradiol 34:E10–E11
Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC (2012) Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 54:555–563
Pope WB, Lai A, Mehta R et al (2011) Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. Am J Neuroradiol 32:882–889
Romano A, Calabria LF, Tavanti F et al (2013) Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol 23:513–520
Saksena S, Jain R, Narang J et al (2010) Predicting survival in glioblastomas using diffusion tensor imaging metrics. J Magn Reson Imaging 32:788–795
Sunwoo L, Choi SH, Park CK et al (2013) Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semiquantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme. J Magn Reson Imaging 37:351–358
Higano S, Yun X, Kumabe T et al (2006) Malignant astrocytic tumors: clinical importance of apparent diffusion coefficient in prediction of grade and prognosis. Radiology 241:839–846
Wangaryattawanich P, Hatami M, Wang J et al (2015) Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression-free survival. Neuro Oncol 17:1525–1537
Ellingson BM, Harris RJ, Woodworth DC et al (2016) Baseline pretreatment contrast enhancing tumor volume including central necrosis is a prognostic factor in recurrent glioblastoma: evidence from single- and multicenter trials. Neuro Oncol. doi:10.1093/neuonc/now187
Zhang M, Gulotta B, Thomas A et al (2016) Large-volume low apparent diffusion coefficient lesions predict poor survival in bevacizumab-treated glioblastoma patients. Neuro Oncol 18:735–743
Gatenby RA, Grove O, Gillies RJ (2013) Quantitative imaging in cancer evolution and ecology. Radiology 269:8–15
Ling CC, Humm J, Larson S et al (2000) Towards multidimensional radiotherapy (MD-CRT): biological imaging and biological conformality. Int J Radiat Oncol Biol Phys 47:551–560
Cui Y, Tha KK, Terasaka S et al (2016) Prognostic imaging biomarkers in glioblastoma: development and independent validation on the basis of multiregion and quantitative analysis of MR images. Radiology 278:546–553
Chang K, Zhang B, Guo X et al (2016) Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol. doi:10.1093/neuonc/now086
Chenevert TL, Malyarenko DI, Newitt D et al (2014) Errors in quantitative image analysis due to platform-dependent image scaling (vol 7, pg 65, 2014). Transl Oncol 7:523
Ellingson BM, Lai A, Nguyen HN, Nghiemphu PL, Pope WB, Cloughesy TF (2015) Quantification of nonenhancing tumor burden in gliomas using effective T-2 maps derived from dual-echo turbo spin-echo MRI. Clin Cancer Res 21:4373–4383
Gevaert O, Mitchell LA, Achrol AS et al (2014) Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273:168–174
Gutman DA, Cooper LA, Hwang SN et al (2013) MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267:560–569
Jamshidi N, Diehn M, Bredel M, Kuo MD (2014) Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology 270:212–222
Zinn PO, Mahajan B, Sathyan P et al (2011) Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One 6:e25451
Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2010) elastix: a toolbox for intensity-based medical image registration. Ieee Trans Med Imaging 29:196–205
Mohammadi S, Moller HE, Kugel H, Muller DK, Deppe M (2010) Correcting eddy current and motion effects by affine whole-brain registrations: evaluation of three-dimensional distortions and comparison with slicewise correction. Magn Reson Med 64:1047–1056
Botev ZI, Grotowski JF, Kroese DP (2010) Kernel density estimation via diffusion. Ann Stat 38:2916–2957
Ogura A, Tamura T, Ozaki M et al (2015) Apparent diffusion coefficient value is not dependent on magnetic resonance systems and field strength under fixed imaging parameters in brain. J Comput Assist Tomogr 39:760–765
Lemkaddem A, Daducci A, Vulliemoz S et al (2012) A multi-center study: intra-scan and inter-scan variability of diffusion spectrum imaging. Neuroimage 62:87–94
Grech-Sollars M, Hales PW, Miyazaki K et al (2015) Multi-centre reproducibility of diffusion MRI parameters for clinical sequences in the brain. NMR Biomed 28:468–485
Barajas RF, Hodgson JG, Chang JS et al (2010) Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology 254:564–576
McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, Trus BL (2001) Medical Image Processing, Analysis & Visualization in clinical research. Fourteenth Ieee Symposium on Computer-Based Medical Systems, Proceedings: 381–386
Goldman M, Craft B, Swatloski T et al (2015) The UCSC cancer genomics browser: update 2015. Nucleic Acids Res 43:D812–D817
Zhu J, Sanborn JZ, Benz S et al (2009) The UCSC cancer genomics browser. Nat Methods 6:239–240
Harrell FE (2001) Regression modeling strategies : with applications to linear models, logistic regression, and survival analysis. Springer, New York
Leijenaar RT, Carvalho S, Velazquez ER et al (2013) Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 52:1391–1397
Parmar C, Rios Velazquez E, Leijenaar R et al (2014) Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 9, e102107
Macyszyn L, Akbari H, Pisapia JM et al (2016) Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 18:417–425
Kickingereder P, Bonekamp D, Nowosielski M et al (2016) Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. doi:10.1148/radiol.2016161382:161382
Cancer Genome Atlas Research N (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068
Engelman JA (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9:550–562
Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred Yung WK (2012) Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol 14:819–829
Sanson M, Marie Y, Paris S et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27:4150–4154
Patel AP, Tirosh I, Trombetta JJ et al (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401
Sottoriva A, Spiteri I, Piccirillo SG et al (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110:4009–4014
Barajas RF Jr, Phillips JJ, Parvataneni R et al (2012) Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol 14:942–954
Hu LS, Ning S, Eschbacher JM et al (2016) Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. doi:10.1093/neuonc/now135
Chu HH, Choi SH, Ryoo I et al (2013) Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology 269:831–840
Park JE, Kim HS, Goh MJ, Kim SJ, Kim JH (2015) Pseudoprogression in patients with glioblastoma: assessment by using volume-weighted voxel-based multiparametric clustering of MR imaging data in an independent test set. Radiology 275:792–802
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Ruijiang Li.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Funding
This research was partially funded by the NIH (grant number: R01 CA193730), and partially supported by the Global Institution for Collaborative Research and Education (GI-CoRE), Hokkaido University, founded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Part of the data used in this research was obtained from The Cancer Imaging Archive (TCIA) sponsored by the Cancer Imaging Program, DCTD/NCI/NIH.
Statistics and biometry
One of the authors has significant statistical expertise.
Ethical approval
Institutional Review Board approval was obtained.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Methodology
• retrospective
• diagnostic or prognostic study
• multicentre study
Rights and permissions
About this article
Cite this article
Cui, Y., Ren, S., Tha, K.K. et al. Volume of high-risk intratumoral subregions at multi-parametric MR imaging predicts overall survival and complements molecular analysis of glioblastoma. Eur Radiol 27, 3583–3592 (2017). https://doi.org/10.1007/s00330-017-4751-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-017-4751-x