Abstract
Key message
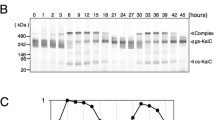
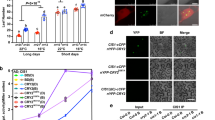
CCA1α and CCA1β protein variants respond to environmental light and temperature cues, and higher temperature promotes CCA1β protein production and causes its retention detectable in the cytoplasm.
Abstract
CIRCADIAN CLOCK ASSOCIATED1 (CCA1), as the core transcription factor of circadian clock, is involved in the regulation of endogenous circadian rhythm in Arabidopsis. Previous studies have shown that CCA1 consists of two abundant splice variants, fully spliced CCA1α and intron-retaining CCA1β. CCA1β is believed to form a nonfunctional heterodimer with CCA1α and its closed-related homolog LHY. Many studies have established that CCA1β is a transcription product, while how CCA1β protein is produced and how two CCA1 isoforms respond to environmental cues have not been elucidated. In this study, we identified CCA1α and CCA1β protein variants under different photoperiods with warm or cold temperature cycles, respectively. Our results showed that CCA1 protein production is regulated by prolonged light exposure and warm temperature. The protein levels of CCA1α and CCA1β peak in the morning, but the detection of CCA1β is dependent on immunoprecipitation enrichment at 22 °C. Higher temperature of 37 °C promotes CCA1β protein production and causes its retention to be detectable in the cytoplasm. Overall, our results indicate that two splice variants of the CCA1 protein respond to environmental light and temperature signals and may, therefore, maintain the circadian rhythms and give individuals the ability to adapt to environment.





Similar content being viewed by others
References
Abdalla KO, Thomson JA, Rafudeen MS (2009) Protocols for nuclei isolation and nuclear protein extraction from the resurrection plant Xerophyta viscosa for proteomic studies. Anal Biochem 384:365–367
Aschoff J (1960) Exogenous and endogenous components in circadian rhythms. Cold Spring Harbor Symp Quant Biol 25:11–28
Boothby TC, Zipper RS, van der Weele CM, Wolniak SM (2013) Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev Cell 24:517–529
Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F (2012) Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet 8:e1002825
Carré IA, Kim J-Y (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53:1551–1557
Cha JY, Kim J, Kim TS, Zeng Q, Wang L, Lee SY, Kim WY, Somers DE (2017) GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat Commun 8:3
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J Cell Mol Biol 16:735–743
Creux N, Harmer S (2019) Circadian rhythms in plants. Cold Spring Harb Perspect Biol 11:a04611
Filichkin SA, Mockler TC (2012) Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol Direct 7:20
Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20:45–58
Filichkin SA, Cumbie JS, Dharmawardhana P, Jaiswal P, Chang JH, Palusa SG, Reddy AS, Megraw M, Mockler TC (2015) Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant 8:207–227
Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283:23073–23083
Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18:1177–1187
Greenham K, McClung CR (2015) Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16:598–610
Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105:4939–4944
Hayama R, Sarid-Krebs L, Richter R, Fernandez V, Jang S, Coupland G (2017) PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J 36:904–918
Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA (2013) Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502:689–692
Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Gonçalves J, Davis SJ (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24:428–443
James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW, Nimmo HG (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24:961–981
Johnson CH, Elliott JA, Foster R (2003) Entrainment of circadian programs. Chronobiol Int 20:741–774
Jones MA, Williams BA, McNicol J, Simpson CG, Brown JW, Harmer SL (2012) Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell 24:4066–4082
Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3:576–583
Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, Dinh HQ, Barta A, Brown JW (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40:2454–2469
Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW (2009) Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21:2045–2057
Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J Cell Mol Biol 88:1058–1070
Lee K, Seo PJ (2018) The HAF2 protein shapes histone acetylation levels of PRR5 and LUX loci in Arabidopsis. Planta 248:513–518
Lei J, Jayaprakasha GK, Singh J, Uckoo R, Borrego EJ, Finlayson S, Kolomiets M, Patil BS, Braam J, Zhu-Salzman K (2019) CIRCADIAN CLOCK-ASSOCIATED1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol 181:1344–1359
Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66(117–128):e115
Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM (2011) The jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol 155:906–915
Lu H, McClung CR, Zhang C (2017) Tick tock: circadian regulation of plant innate immunity. Annu Rev Phytopathol 55(287):311
Ma Y, Gil S, Grasser KD, Mas P (2018) Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in Arabidopsis. Plant Cell 30:907–924
Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22:1184–1195
Marshall CM, Tartaglio V, Duarte M, Harmon FG (2016) The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28:2560–2575
McClung CR (2019) The plant circadian oscillator. Biology 8:14
Meier I, Somers DE (2011) Regulation of nucleocytoplasmic trafficking in plants. Curr Opin Plant Biol 14:538–546
Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2:629–641
Mwimba M, Karapetyan S, Liu L, Marques J, McGinnis EM, Buchler NE, Dong X (2018) Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat Commun 9:4290
Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46:686–698
Nicolas M, Rodriguez-Buey ML, Franco-Zorrilla JM, Cubas P (2015) A recently evolved alternative splice site in the BRANCHED1a Gene controls potato plant architecture. Curr Biol CB 25:1799–1809
Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23:1061–1069
Panter PE, Muranaka T, Cuitun-Coronado D, Graham CA, Yochikawa A, Kudoh H, Dodd AN (2019) Circadian regulation of the plant transcriptome under natural conditions. Front Genet 10:1239
Perales M, Mas P (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19:2111–2123
Perez-Santangelo S, Mancini E, Francey LJ, Schlaen RG, Chernomoretz A, Hogenesch JB, Yanovsky MJ (2014) Role for LSM genes in the regulation of circadian rhythms. Proc Natl Acad Sci USA 111:15166–15171
Pittendrigh CS, Minis DH (1964) The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat 98:261–322
Salmela MJ, Weinig C (2019) The fitness benefits of genetic variation in circadian clock regulation. Curr Opin Plant Biol 49:86–93
Salomé PA, McClung CR (2005a) What makes the Arabidopsis clock tick on time? A review on entrainment. Plant Cell Environ 28:21–38
Salomé PA, McClung CR (2005b) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17:791–803
Salomé PA, Oliva M, Weigel D, Krämer U (2013) Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO J 32:511–523
Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, Brown JW, Cerdan PD, Borevitz JO, Mas P, Ceriani MF, Kornblihtt AR, Yanovsky MJ (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468:112–116
Schlaen RG, Mancini E, Sanchez SE, Perez-Santangelo S, Rugnone ML, Simpson CG, Brown JW, Zhang X, Chernomoretz A, Yanovsky MJ (2015) The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc Natl Acad Sci USA 112:9382–9387
Seo PJ, Hong SY, Kim SG, Park CM (2011a) Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci 16:541–549
Seo PJ, Kim MJ, Ryu JY, Jeong EY, Park CM (2011b) Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat Commun 2:303
Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24:2427–2442
Shakhmantsir I, Nayak S, Grant GR, Sehgal A (2018) Spliceosome factors target timeless (tim) mRNA to control clock protein accumulation and circadian behavior in Drosophila. eLife 7:e39821
Shi G, Xie P, Qu Z, Zhang Z, Dong Z, An Y, Xing L, Liu Z, Dong Y, Xu G, Yang L, Liu Y, Xu Y (2016) Distinct roles of HDAC3 in the core circadian negative feedback loop are critical for clock function. Cell Rep 14:823–834
Somers DE, Devlin P, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–1490
Song HR, Noh YS (2012) Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol Cells 34:279–287
Song Q, Huang TY, Yu HH, Ando A, Mas P, Ha M, Chen ZJ (2019) Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol 20:170
Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW (2012) Alternative splicing in plants–coming of age. Trends Plant Sci 17:616–623
Tong M, Lee K, Ezer D, Cortijo S, Jung J, Charoensawan V, Box MS, Jaeger KE, Takahashi N, Mas P, Wigge PA, Seo PJ (2020) The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol 182:612–625
Wang L, Fujiwara S, Somers DE (2010a) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29:1903–1915
Wang L, Fujiwara S, Somers DE (2010b) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29:1903–1915
Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, Jiao Y, Puta F, McClung CR, Xu X, Ma L (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24:3278–3295
Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110:761–766
Wang Y, He Y, Su C, Zentella R, Sun TP, Wang L (2020) Nuclear localized O-fucosyltransferase SPY facilitates PRR5 proteolysis to fine-tune the pace of Arabidopsis circadian clock. Mol Plant 13:446–458
Webb AAR, Seki M, Satake A, Caldana C (2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10:550
Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, Zhao C, McClung CR, Xu X (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26:2843–2857
Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM (2009a) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150:844–857
Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM (2009b) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150:844–857
Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32:617–630
Acknowledgements
This work was supported by the National Natural Science Foundation of China to S.Z. (31700255), Q.X. (31670285), and X.X. (U1904202), the Postdoctoral Science Foundation of China (2017M621098), the Natural Science Foundation of Hebei to Q.X. (17966304D) and S.Z. (C2017205134), and the Hebei Hundred Talents Program (E2016100018) to Q.X.
Author information
Authors and Affiliations
Contributions
QX, XX and SZ conceived the project and wrote the article. S.Z. performed most of the experiments. SZ and HL completed the qRT-PCR experiment; SZ, XL, and LW completed vector construction and transgenic work; SZ and LY completed the hypocotyl length measurement, SZ, LY, QX and XX analyzed the data.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Da-Bing Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession numbers
Sequence data for the genes referred in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CCA1, AT2G46830; IPP2, At3g02780.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, S., Liu, H., Yuan, L. et al. Recognition of CCA1 alternative protein isoforms during temperature acclimation. Plant Cell Rep 40, 421–432 (2021). https://doi.org/10.1007/s00299-020-02644-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02644-7