Abstract
Key message
Lily R3-MYB transcription factors are involved in negative regulation to limit anthocyanin accumulation in lily flowers and leaves and create notable color patterns on ectopically expressed petunia flowers.
Abstract
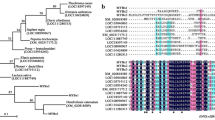
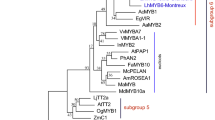
In eudicots, both positive and negative regulators act to precisely regulate the level of anthocyanin accumulation. The R3-MYB transcription factor is among the main factors repressing anthocyanin biosynthesis. Although, in monocots, the positive regulators have been well characterized, the negative regulators have not been examined. Two R3-MYBs, LhR3MYB1 and LhR3MYB2, which were identified in lily transcriptomes, were characterized in this study to understand the regulatory mechanisms of anthocyanin biosynthesis. LhR3MYB1 and LhR3MYB2 had a C2 suppressor motif downstream of a single MYB repeat; the similar amino acid motif appears only in AtMYBL2 among the eudicot R3-MYB proteins. Stable and transient overexpression of LhR3MYB1 and LhR3MYB2 in tobacco plants showed suppression of anthocyanin biosynthesis by both; however, suppression by LhR3MYB2 was stronger than that by LhR3MYB1. In the lily plant, the LhR3MYB2 transcript was detected in leaves with light stimulus-induced anthocyanin accumulation and in pink tepals. Although LhR3MYB1 was expressed in some, but not all tepals, its expression was not linked to anthocyanin accumulation. In addition, LhR3MYB1 expression levels in the leaves remained unchanged by the light stimulus, and LhR3MYB1 transcripts predominantly accumulated in the ovaries, which did not accumulate anthocyanins. Thus, although LhR3MYB1 and LhR3MYB2 have an ability to repress anthocyanin accumulation, LhR3MYB2 is more strongly involved in the negative regulation to limit the accumulation than that by LhR3MYB1. In addition, the overexpression of LhR3MYB2 generated notable color patterns in petunia flowers; thus, the usefulness of the LhR3MYB genes for creating unique color patterns by genetic engineering is discussed.






Similar content being viewed by others
References
Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28:319–332
Albert NW, Lewis DH, Zhang H, Irving LJ, Jameson PE, Davies KM (2009) Light-induced vegetative anthocyanin pigmentation in Petunia. J Exp Bot 60:2191–2202
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014a) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980
Albert NW, Davies KM, Schwinn KE (2014b) Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal Behav 9:e29526
Azadi P, Bagheri H, Nalousi AM, Nazari F, Chandler SF (2016) Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol Adv 34:1073–1090
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107:467–590
Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167:1448–1470
Chu Y, Pan J, Wu A, Cai R, Chen H (2014) Molecular cloning and functional characterization of dihydroflavonol-4-reductase gene from Calibrachoa hybrida. Sci Hortic 165:398–403
Colanero S, Perata P, Gonzali S (2018) The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front Plant Sci 9:830
Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG (2011) PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H. J Exp Bot 62:1133–1143
Comber HF (1949) A new classification of the genus Lilium. In: Lily yearbook, vol 13. The Royal Horticultural Society, London, pp 85–105
Cui L, Shan J, Shi M, Gao J, Lin H (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Dubos C, Le GJ, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul J-M, Alessandro A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Elomaa P, Helariutta Y, Kotilainen M, Teeri TH, Griesbach RJ, Seppänen P (1995) Transgene inactivation in Petunia hybrida is influenced by the properties of the foreign gene. Mol Gen Gen 248:649–656
Gates DJ, Olson BJSC, Clemente TE, Smith SD (2018) A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol 217:1346–1356
Glover BJ, Martin C (2012) Anthocyanins. Curr Biol 22:R147–R150
Gould KS (2004) Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004:314–320
Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Hsu YH, Tagami T, Matsunaga K, Okuyama M, Suzuki T, Noda N, Suzuki M, Shimura H (2017) Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus. Plant J 89:325–337
Hughes NM, Neufeld HS, Burkey KO (2005) Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol 168:575–587
Jeknić Z, Morré JT, Jeknić S, Jevremović S, Subotić A, Chen THH (2012) Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from tiger lily (Lilium lancifolium Thunb. ‘Splendens’). Plant Cell Physiol 53:1899–1912
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Noriko N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao G, Nehra NS, Lu C, Dyson BK, Tsuda S, Ashikari T, Kusumi T, Mason JG, Tanaka Y (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48:1589–1600
Koseki M, Goto K, Masuta C, Kanazawa A (2005) The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol 46:1879–1883
Lai Y, Shimoyamada Y, Nakayama M, Yamagishi M (2012) Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci 193–194:136–147
Lai Y, Li H, Yamagishi M (2013) A review of target gene specificity of flavonoid R2R3-MYB transcription factors and a discussion of factors contributing to the target gene selectivity. Front Biol 8:577–598
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X Version 2.0. Bioinformatics 23:2947–2948
Leslie AC (1982) The international lily register, 3rd edn. The Royal Horticultural Society, London
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Marasek-Ciolakowska A, Nishikawa T, Shea DJ, Keiichi Okazaki K (2018) Breeding of lilies and tulips—interspecific hybridization and genetic background. Breed Sci 68:35–52
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Matus JT, Cavallini E, Loyola R, Höll J, Finezzo L, Santo SD, Vialet S, Commisso M, Roman F, Schubert A, Alcalde JA, Bogs J, Ageorges A, Tornielli GB, Arce-Johnson P (2017) A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J 91:220–236
Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H (1992) Endogenous and environmental factors influence 35S promoter methylation of a maize Al gene construct in transgenic petunia and its colour phenotype. Mol Gen Genet 231:345–352
Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M (2012) Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J 70:739–749
Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N (2009) Light-induced expression of basic helix–loop–helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic 121:84–91
Nakatsuka T, Yamada E, Saito M, Fujita K, Nishihara M (2013) Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep 32:1925–1937
Nakayama M (2014) A research strategy to understand the mechanisms that govern flower color pattern formation. JARQ Jpn Agric Res Q 48:271–277
Nemie-Feyissa D, Olafsdottir SM, Heidari B, Lillo C (2014) Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98:34–40
Noda N, Yoshioka S, Kishimoto S, Nakayama M, Douzono M, Tanaka Y, Aida R (2017) Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci Adv 3:e1602785
Noman A, Aqeel M, Deng J, Khalid N, Sanaullah T, Shuilin H (2017) Biotechnological advancements for improving floral attributes in ornamental plants. Front Plant Sci 8:530
Nørbæk R, Kondo T (1999) Anthocyanins from flowers of Lilium (Liliaceae). Phytochem 50:1181–1184
Oud JSN, Schneiders H, Kool AJ, van Grinsven MQJM (1995) Breeding of transgenic orange Petunia hybrida varieties. Euphytica 85:403–409
Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L (2010) Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231:1061–1076
Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18:1274–1291
Schwinn KE, Ngo H, Kenel F, Brummell DA, Albert NW, McCallum JA, Pither-Joyce M, Crowhurst RN, Eady C, Davies KM (2016) The Onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front Plant Sci 7:1865
Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311:566–578
Spelt C, Quattrocchio F, Mol J, Koes R (2000) anthocyanin1 of petunia encodes a basic-helix loop helix protein that directly activates structural anthocyanin genes. Plant Cell 12:1619–1631
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Suzuki K, Tasaki K, Yamagishi M (2015) Two distinct spontaneous mutations involved in white flower development in Lilium speciosum. Mol Breed 35:193
Suzuki K, Suzuki T, Nakatsuka T, Dohra H, Yamagishi M, Matsuyama K, Matsuura H (2016) RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom 17:611
Tanaka Y, Ohmiya A (2008) Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotechnol 19:190–197
Tsuda S, Fukui Y, Nakamura N, Katsumoto Y, Yonekura-Sakakibara K, Fukuchi-Mizutani K, Ohira K, Ueyama Y, Ohkawa H, Holton TA, Kusumi T, Tanaka T (2004) Flower color modification of Petunia hybrida commercial varieties by metabolic engineering. Plant Biotech 21:377–386
Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17(5):1612–1624
Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5:133
Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen J (2008) Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol 8:81
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci 20:176–185
Yamagishi M (2011) Oriental hybrid lily Sorbonne homologue of LhMYB12 regulates anthocyanin biosyntheses in flower tepals and tepal spots. Mol Breed 28:381–389
Yamagishi M (2013) How genes paint lily flowers: regulation of colouration and pigmentation patterning. Sci Hortic 163:27–36
Yamagishi M (2016) A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol Breed 36:3
Yamagishi M (2018) Involvement of a LhMYB18 transcription factor in large anthocyanin spot formation on the flower tepals of the Asiatic hybrid lily (Lilium spp.) cultivar ‘Grand Cru’. Mol Breed 38:60
Yamagishi M, Kishimoto S, Nakayama M (2010a) Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed 129:100–107
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010b) Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol 51:463–474
Yamagishi M, Yoshida Y, Nakayama M (2012) The transcription factor LhMYB12 determines anthocyanin pigmentation in the tepals of Asiatic hybrid lilies (Lilium spp.) and regulates pigment quantity. Mol Breed 30:913–925
Yamagishi M, Toda S, Tasaki K (2014) The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol 201:1009–1020
Yamagishi M, Uchiyama H, Handa T (2018) Floral pigmentation pattern in Oriental hybrid lily (Lilium spp.) cultivar ‘Dizzy’ is caused by transcriptional regulation of anthocyanin biosynthesis genes. J Plant Physiol 228:85–91
Yoshida K, Oyama-Okubo N, Yamagishi M (2018) An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Mol Breed 38:144
Yuan Y-W, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD Jr. (2013) Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194:255–263
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Acknowledgements
This work was supported by a Grant-In-Aid for Scientific Research (No. 15H04447) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sakai, M., Yamagishi, M. & Matsuyama, K. Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep 38, 609–622 (2019). https://doi.org/10.1007/s00299-019-02391-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02391-4