Abstract
Fusarium pseudograminearum is an important pathogen of Fusarium crown rot and Fusarium head blight, which is able to infect wheat and barley worldwide, causing great economic losses. Transcription factors (TFs) of the basic leucine zipper (bZIP) protein family control important processes in all eukaryotes. In this study, we identified a gene, designated FpAda1, encoding a bZIP TF in F. pseudograminearum. The homolog of FpAda1 is also known to affect hyphal growth in Neurospora crassa. Deletion of FpAda1 in F. pseudograminearum resulted in defects in hyphal growth, mycelial branching and conidia formation. Pathogenicity assays showed that virulence of the Δfpada1 mutant was dramatically decreased on wheat coleoptiles and barley leaves. However, wheat coleoptile inoculation assay showed that Δfpada1 could penetrate and proliferate in wheat cells. Moreover, the FpAda1 was required for abnormal nuclear morphology in conidia and transcription of FpCdc2 and FpCdc42. Taken together, these results indicate that FpAda1 is an important transcription factor involved in growth and development in F. pseudograminearum.
Similar content being viewed by others
Introduction
The plant pathogen Fusarium pseudograminearum is the causative agent of Fusarium crown rot (FCR) in wheat and barley, resulting in substantial yield losses worldwide (Kazan and Gardiner 2018). Particularly, in the Huanghuai wheat-growing region of China, it has been reported that F. pseudograminearum was the dominant pathogen of FCR (Li et al. 2012; Zhou et al. 2019). F. pseudograminearum was initially recognized as a population within the Fusarium graminearum species group (group 1). However, F. pseudograminearum is heterothallic and it was segregated by molecular analyses (Aoki and O’Donnell 1999; Gardiner et al. 2018). Like F. graminearum, F. pseudograminearum also causes Fusarium head blight (FHB) and produces deoxynivalenol (DON) mycotoxin under favorable conditions (Kazan and Gardiner 2018; Obanor et al. 2013). Despite the devastating effects caused by FCR and FHB, establishing effective disease management strategies has been very difficult. Therefore, understanding the molecular mechanism of pathogenicity in F. pseudograminearum is of utmost relevance, given its value in the design of a proper strategy for FCR and FHB disease management.
Transcription factors (TFs) are DNA-binding proteins that interact with other components of the transcriptional machinery to regulate the expression of multiple genes. TFs can be classified into several categories based on primary and/or three-dimensional structure similarities in the DNA-binding and multimerization domains (Riechmann et al. 2000; Warren 2002). The family of transcription factors containing a basic leucine zipper domain (bZIP) is widely distributed across eukaryotes (Hurst 1995; Kong et al. 2015). In plants, bZIP proteins are the largest protein family, which regulate processes including abiotic stress, seed maturation, flower development and pathogen defense (Alves et al. 2013; Amorim et al. 2017). In Saccharomyces cerevisiae, the bZIP TF family contains 14 genes, and the largest is the YAP1 group, formed by eight members. Five YAP1 family members (YAPs 1, 2, 4, 5 and 6) have been implicated in oxidative stress and DNA-damage responses (He and Fassler 2005; Tan et al. 2008; Workman et al. 2006). Filamentous fungi typically contain well over a dozen of these TFs. Several fungal bZIPs have been characterized and implicated in multiple phenomena including remediation of development, amino acid biosynthesis, unfolded protein response, nutrient utilization and various stress responses (Guo et al. 2010; Kong et al. 2015; Son et al. 2011). The bZIP protein AP-1 is essential for pathogens’ growth, development, infection and pathogenicity in Magnaporthe oryzae, Ustilago maydis and Colletotrichum gloeosporioides, among others (Guo et al. 2011; Li et al. 2017; Molina and Kahmann, 2007). In N. crassa, out of nine characterized bZIP members, Ada-1 (all development altered-1) is unique by regulating growth under minimal media conditions (Colot et al. 2006; Tian et al. 2011). In F. graminearum, a total of 22 bZIP TFs were functionally analyzed, and six TFs were associated with growth and pathogenicity. Among these, deletion of an Ada-1 homolog (GzbZIP001) resulted in growth and virulence defects in F. graminearum (Son et al. 2011). However, there has been no research on Ada-1-like transcription factor in F. pseudograminearum, and their regulatory mechanism is not clear.
Cell cycle regulation is pivotal for proper cell division and cellular differentiation in eukaryotic cells. The central regulators that govern eukaryotic cell cycle progression are cyclin-dependent kinases and their partners (Bloom and Cross 2007; Humphrey and Pearce 2005; Sendinc et al. 2015). In model organisms such as yeast and N. crassa, Cdc2 is essential for cell cycle progression and hyphal growth (Booher and Beach 1986; Borkovich et al. 2004). F. graminearum has two Cdc2 genes, Cdc2A and Cdc2B. The two Cdc2 orthologs have reproduction functions in hyphal growth and asexual reproduction, and only Cdc2A is important for plant infection and sexual reproduction (Jiang et al. 2016; Liu et al. 2015). Cdc42p is a Rho family GTPase, required for changes in polarized growth during mating and pseudohyphal development in S. cerevisiae. Cdc42p homologs in higher organisms are also associated with changes in cell shape and polarity (Moran et al. 2019; Rincon et al. 2014). The Cdc42 homolog has also been found in many fungi strains, and is required for hyphal growth (Bassilana et al. 2005; Boyce et al. 2001).
In this study, we identified FpAda1 as a homolog of the bZIP transcription factor Ada-1 in F. pseudograminearum, which was found to be involved in hyphal growth, conidiation and pathogenicity. Nuclear formation and the expression of cyclin-dependent protein kinase genes in Δfpada1 was also studied to understand its possible regulatory network.
Materials and methods
Sequence analysis of FpAda1
The Ada-1 (all development altered-1) gene (locus NCU00499) of N. crassa was downloaded from NCBI and used as the query to search against the F. pseudograminearum genome by BlastP and tBlastN algorithms (Altschul et al. 1990; Gardiner et al. 2018). The b-ZIP domain of FpAda1 was predicted by SMART (http://smart.emblheidelberg.de).
qRT-PCR analyses
For total RNA extraction, conidia were induced in CMC medium at 150 rpm, 25 °C in the dark for 4 days. Mycelia were obtained by cultivating conidia with YEPD liquid medium at 25 °C, 150 rpm for 12 h and were then harvested by filtration over two layers of miracloth and washed with sterilized water. For conidial infection (IF18 h to IF7 days), wheat cultivar Aikang 58, which is susceptible to F. pseudograminearum, was grown in a greenhouse at 25 °C for 4 days. Two milliliters of conidia suspension (1 × 107/ml) was infected on each coleoptile of wheat seedlings. After 18 h, 30 h, 2 days, 3 days, 5 days and 7 days’ incubation in dark at 25 °C, lesion areas with 5 mm extension were harvested. Total RNA was extracted from each sample with the RNAsimple Total RNA Kit (Tiangen, China) according to the manufacturer’s protocol. RNA was further purified and cDNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, China).
The expression levels of FpAda1 and tested FpCdc2, FpCdc25, FpCdc42 and FpBub1 were determined by quantitative real-time PCR (qRT-PCR) using the primers listed in Supplementary Table S1. For each sample, the FpTEF1 gene was used as an internal control, and the following conditions were used for the qRT-PCR reaction: 95 °C for 30 s, 40 cycles at 95 °C for 5 s and 60 °C for 31 s to calculate cycle threshold values, followed by a dissociation program of 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s to obtain melt curves. The transcript levels of test genes were determined according to the function ΔCT = CT (test gene)—CT (reference gene). To compare untreated and treated expression levels, the function ΔΔCT was determined using the equation ΔΔCT = ΔCT (treatment) − ΔCT (control) where the control was mock-treated with F. pseudograminearum mycelia. The induction ratio of treatment/control was then calculated 2−ΔΔCT.
Generation of the FpAda1 deletion mutant and complementation strains
The split-marker approach was used to generate gene-replacement constructs for the FpAda1 gene as described in our previous study (Chen et al. 2019a). Primers are listed in Supplementary Table S1 and a schematic diagram of primers located for gene replacement with split-marker strategy and screening of mutant is shown in Fig. 2a. Briefly, the 1147-bp upstream and 1125-bp downstream flanking sequences were amplified with primer pairs F1/R1 and F2/R2, respectively. The hygromycin gene (hph) was amplified from pkov21 with primer pairs HYG/F and HYG/R. After three PCR cycles, a 1911-bp fusion PCR product including 5′-flanking region and 5′-hph region was obtained by overlap PCR amplification with primer pair A1 + HY/R using mixed fragments of FpAda1 upstream and hph fragments as templates. At the same time, a 2188-bp fusion PCR product including 3′-hph region and 3′-flanking region was obtained by overlap PCR amplification with primer pair YG/F + B2 using mixed fragments of FpAda1 downstream and hph fragments as templates. Products obtained by the third PCR cycle were used for fungal transformation. Putative gene deletion mutants were identified by PCR assays using the primers G1/G2, H2F/H2R, F3/H1R and H1F/R3. Genome DNA was digested by EcoR I and separated by agarose gel electrophoresis. The hygromycin gene was detected by the DIG DNA Labeling and Detection Kit (Roche, USA) according to the manufacturer’s protocol.
The plasmid pYIP-102 was used for construction of the complementation vector. The FpAda1 gene with its native promoter was amplified using primers ComF/ComR and inserted into the vector. The constructed vector was transformed into a Δfpada1 mutant. The complemented transformants were confirmed by western blot analysis.
Phenotype determination
For mycelial growth assays, 5-mm mycelial plugs were taken from the edge of a 3-day-old colony of each strain and placed on PDA plates and incubated at 25 °C. Mycelial morphology was observed 12 h later, and colony diameters were measured and photographed 3 days later. For the conidiation assay, two 5-mm plugs from the edge of a 3-day-old colony of each strain were inoculated in 100 ml CMC. After 4 days’ cultivation in a 150-rpm-shaker at 25 °C, conidia were harvested by filtering through a layer of miracloth and counted using a hemocytometer. For the conidia germination assay, 0.1 ml of 104 conidia/ml suspension was prepared and cultured in sterile distilled water at 25 °C in the dark for 3 h and 6 h. Three biological replicates were used for each strain and each experiment was repeated three times independently. Data were analyzed using a Student’s t test. To probe for nuclei, 2 µM DAPI dilactate (Takara, China) was used.
Pathogenicity assays
For virulence on wheat coleoptiles, 5-mm mycelial plugs from the edge of a 3-day-old PDA plate of each strain were inoculated onto the wheat coleoptiles of the susceptible cultivar Aikang 58. The fungal discs were removed after 24 h, and seedling lesion lengths were photographed at 3 days post-inoculation (dpi). All experiments were performed three times with five replicates per experiment. For virulence on malting barley leaves, barley seeds were planted in pots for 14 days, and then 5-mm mycelial plugs from the edge of a 3-day-old PDA plate of each strain were inoculated onto the barley leaves. All experiments were performed three times with five replicates per experiment. For pot-culture experiments, susceptible wheat cultivar Aikang 58 plants were planted in sterile soil mixing 0.5% inoculation millet for 10 days. Then wheat growth and infection were analyzed and documented. For observation of mycelium growth in infected coleoptiles, the inoculated wheat coleoptiles were harvested after 30 h, and epidermal cells were viewed under a Nikon Ti-s instrument.
Results
Identification and expression of FpAda1 in F. pseudograminearum
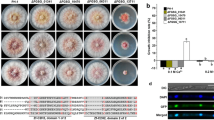
One putative all development altered-1 gene (FPSE_04421, designated as FpAda1) in F. pseudograminearum was retrieved by BLAST search of the F. pseudograminearum genome with the N. crassa Ada-1 (NCU00499) as a query. The FpAda1 gene is predicted to encode a 598-amino acid protein showing 61% identity match to N. crassa Ada-1. The domain analysis showed that FpAda1 has a conserved bZIP DNA-binding domain (Fig. 1a).
Sequence alignment and expression profiles of FpAda1. a Sequence alignment of the predicted amino acid sequence of FpAda1 with its ortholog from Neurospora crassa (NcAda-1). The red box indicates the b-ZIP domain. b Expression of FpAda1 in hyphae, conidia, and infected wheat coleoptiles from 18 h to 7 days post fertilization
To further investigate the potential functions of FpAda1 gene during development and pathogenicity in F. pseudograminearum, total RNA samples of mycelia, conidia and conidial infection wheat plants (IF18 h to IF7 days) were obtained. By qRT-PCR we observed that FpAda1 expression was induced during conidiation and early infection stages (IF18 h and IF30 h), and a high transcriptional level of FpAda1 was also detected at IF5 days (Fig. 1b). These results indicate that FpAda1 might play roles in both conidiation and virulence.
Deletion and complementation of FpAda1 gene in F. pseudograminearum
To determine the biological function of FpAda1 gene in F. pseudograminearum, FpAda1 deletion mutants were generated. In Fig. 2a, a schematic diagram shows the strategy that was used to generate FpAda1 gene deletion mutants and molecular verification of Δfpada1. Transformants were selected on hygromycin-amended medium, and four individual targeted deletion mutants, designated Δfpada1-T1, Δfpada1-T2, Δfpada1-T3 and Δfpada1-T4, were created and checked by PCR (Fig. 2b). However, due to bacterial contaminations, Δfpada1-T4 was discarded. Finally, Δfpada1-T1 and Δfpada1-T3 were confirmed as FpAda1 gene knock-out transformants by southern blot analysis (Fig. 2c). In order to confirm that phenotypic defects in mutants were caused by FpAda1 gene deletion, we complemented the mutant with a wild-type FpAda1 gene with its native promoter, and a FLAG-tag was fused to the C-terminal of FpAda1. We confirmed the complemented strain Δfpada1-C by western blot (Fig. S1).
Generation and identification of FpAda1 gene deletion mutant. a Gene deletion strategy for FpAda1. Primers used for gene replacement and screening of mutant are indicated by arrows. b Confirmation of FpAda1 deletion mutants by PCR strategy. Verification of incorporation into genomic DNA by PCR using four pairs of primers, which were used to analyze hygromycin (H2F/H2R), upstream (F3/H1R), downstream (H1F/R3) and the FpAda1 gene (G1/G2) positivity. Amplified fragments were 523, 991, 939 and 511-bp long. WT wild-type strain WZ2-8, M molecular markers, H hygromycin gene, F upstream, R downstream, G FpAda1 gene. c Southern blot analysis of WT and Δfpada1 using the 750-bp DNA fragment of hygromycin as a probe. The genomic DNA preparation of each strain was digested with EcoR I
FpAda1 is critical for vegetative growth in F. pseudograminearum
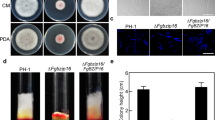
To evaluate the influence of FpAda1 in the vegetative growth of F. pseudograminearum, we examined the growth of Δfpada1 cultured on PDA medium for 3 days. Growth assessment records showed that FpAda1 deletion caused a significant reduction in the strain’s vegetative growth (Fig. 3a, c). In comparison with the wild-type and Δfpada1-C strains, colony pigment deposition increased in Δfpada1 (Fig. 3a). Further microscopic examination showed that the hyphae from the Δfpada1 mutant were thinner and produced less branches at the hyphal tip as compared with the growing hyphae of the wild-type and the complemented strains (Fig. 3b). Thus, FpAda1 played an important role in the growth and hyphal branching in F. pseudograminearum.
Hyphal growth and pigment formation of the Δfpada1 mutant. a The WT, Δfpada1 mutants and the complemented strain Δfpada1-C were grown on PDA plates for 3 days. b Colony diameters were assayed. Linear bars in each column denote standard errors of three experiments. Two asterisks indicate significant difference of colony diameter (P < 0.01). c Hyphal tip growth and branching patterns of F. pseudograminearum grown on PDA medium for 12 h. Bars = 20 μm
FpAda1 is important for conidiation in F. pseudograminearum
We performed conidial production test in F. pseudograminearum to study the function of FpAda1. Cultures of the Δfpada1 mutant produced few conidia on CMC medium when compared with wild-type and Δfpada1-C strains (Fig. 4a). After 4 days of incubation, only 5.36 ± 0.88 × 105 conidia/ml were obtained from the Δfpada1 mutant, in contrast to 10.27 ± 0.81 × 105 and 10.45 ± 1.30 × 105 conidia/ml in WT and Δfpada1-C strains, respectively (Fig. 4a). These results indicate that FgAda1 is important for conidia production in F. pseudograminearum.
Conidial production of the Δfpada1 mutant. a Conidial production (4 days after incubation in liquid CMC) of WT, Δfpada1 mutants and the complemented strain Δfpada1-C were examined by microscopy. b Number of conidia produced by each line was measured at 4 dai. Data shown are representative of three separate experiments. The bars indicate standard error. **P < 0.01 (t test)
To further study the function of FpAda1 in the development of conidiation, we monitored conidia germination in the Δfpada1 mutant. The conidia collected from WT, Δfpada1 and Δfpada1-C were incubated in sterile distilled water. After 3 h, the conidia in the mutant were able to germinate and we saw a synchronization compared to the wild-type and Δfpada1-C strains (Fig. 5a). However, the tube length of Δfpada1 was obviously shorter than that of WT and Δfpada1-C strains after 6 h (Fig. 5b), which might be a consequence of a reduction in growth rate (Fig. 3a). These results indicated that FpAda1 played important roles in growth and conidiation, but not in conidia germination.
Conidial germination of the Δfpada1 mutant. a Conidia germination rates were measured at 3 h after incubation in three independent biological replicates, each of which comprised of at least five glass slides. The bars indicate standard errors. b Conidia germlings (6 h after incubation in water) were examined by microscopy. Bars = 20 μm
FpAda1 affects the pathogenicity in F. pseudograminearum
To investigate the role of FpAda1 in fungal virulence, we first inoculated wheat coleoptiles with WT, Δfpada1 and Δfpada1-C strains. The average length of brown lesions on the wheat coleoptiles infected with the Δfpada1 mutant was 0.62 ± 0.12 cm, whereas those infected with the WT and Δfpada1-C strains showed average lesion lengths of 1.13 ± 0.15 and 1.06 ± 0.16 cm, respectively (Fig. 6a, b). Furthermore, we also inoculated the aforementioned strains on barley leaves. The Δfpada1 mutant also caused lesions at the inoculated leaves, but the mutant-caused disease effects were less pronounced as compared with the WT and Δfpada1-C under the same conditions (Fig. 6c). Finally, a pot-culture experiment was used to further confirm the involvement of FpAda1 in fungal virulence. The WT and Δfpada1-C caused crown rot symptoms in wheats at 10 days post inoculation (dpi). However, wheat seedlings showed mild symptoms after inoculation with the Δfpada1 mutant (Fig. 6d).
Pathogenicity assays of the Δfpada1 mutant. a Wheat seedling hypocotyls were inoculated with mycelial plugs of WT, Δfpada1 mutants and the complemented strain Δfpada1-C and examined 3 days post-inoculation. b Lesion lengths on wheat hypocotyls were measured at 3 days post-inoculation. The bars indicate the standard errors. **P < 0.01 (t test). c Barley leaves were inoculated with mycelial plugs and examined 5 days post-inoculation. d Wheat growth and wheat root lesions were examined from the pot-culture experiment at 10 days post-inoculation. e Infection mycelia in wheat hypocotyl cells were examined at 30 h post-inoculation. Bars = 20 μm
We then evaluated the effects of Δfpada1 mutant on the fungal invasion process in wheat at a cellular level. After inoculation of wheat coleoptiles, microscopic analysis showed that hyphae of WT, Δfpada1 and Δfpada1-C infected and extended similarly in coleoptile cells (Fig. 6e). The results suggest that the observed reduction in virulence might be a consequence of a reduction in growth rate of Δfpada1 mutant.
FpAda1 is involved in the cell cycle of F. pseudograminearum
Cell cycle regulation has been shown to be important for growth and morphological changes. Because Δfpada1 exhibited severe defects in growth and development, we assessed the expression levels of nucleus and cell division cycle genes. DAPI staining revealed that the Δfpada1 cells had abnormal nuclear morphology in conidia (Fig. 7a). In WT and Δfpada1-C strains, one disc-shaped nucleus could be observed in every cell. However, nuclei-lacking cells were widespread in Δfpada1 conidia. In addition, the expression of three cyclin-dependent protein kinase genes (FpCdc2, FpCdc25 and FpCdc42), involved in fungal growth, were analyzed in WT, Δfpada1 and Δfpada1-C strains. A serine/threonine protein kinase (FpBub1) was chosen as contrast for the cyclin-dependent protein kinase genes. The expression levels of FpCdc2 and FpCdc42 were significantly reduced in Δfpada1 mutant (Fig. 7b), which further supported the role of FpAda1 in cell cycle regulation.
Nuclei of WT, Δfpada1 mutants and the complemented strain Δfpada1-C were stained with DAPI and images were taken. Bars = 10 μm. b Relative transcription levels of cyclin-dependent protein kinase genes (FpCdc2, FpCdc25, FpCdc42) and serine/threonine protein kinase (FpBub1) gene in WT, Δfpada1 mutant and the complemented strain Δfpada1-C. For each gene, the expression level in WT was arbitrarily set as 1. Bars denote standard errors from three repeated experiments. *P < 0.05 (t test)
Discussion
The bZIP transcription factors have been reported to regulate many central physiological and developmental processes in plants, such as flowering, seed maturation, stress response and pathogen defense (Alves et al. 2013; Banerjee and Roychoudhury 2017). Recently, a number of bZIP transcription factors have been identified in plant pathogenic fungi and played important roles in development, stress response and virulence. In M. oryzae, 22 bZIP transcription factors were identified and characterized as being involved in development, nutrient utilization and various stress responses (Kong et al. 2015). For example, MpAtf1 regulated the transcription of laccases and peroxidases, which was critical in pathogenicity (Guo et al. 2010). In F. graminearum, transcription factors related to growth, development, stress responses and virulence were reported (Chen et al. 2019b; Lv et al. 2019). However, few transcription factors have been described in F. pseudograminearum.
In this study, we have characterized a bZIP-type transcription factor FpAda1 in F. pseudograminearum as a homolog of N. crassa Ada-1 protein. Similar to other bZIP proteins, FpAda1 contains a bZIP DNA-binding domain. In N. crassa, the bZIP TF family contains nine genes and can be divided into two groups. Ada-1 was clustered to the GCN4 clade, and the Δada-1 mutant showed a reduced growth rate and very short aerial hyphae. Among the strain carrying a deletion of the bZIP gene, Δada-1 showed the greatest number of expression differences from the WT with 290 genes increasing, and 219 genes decreasing, consistent with its growth defect (Tian et al. 2011).
To determine the biological function of FpAda1 in F. pseudograminearum, its deleted mutant was generated. Compared to wild type, the mutant showed defects in hyphal growth, mycelial branching and conidia formation. However, in F. graminearum, the Ada-1 homolog GzbZIP001 mutant showed no significant changes in conidiation. GzbZIP001 was required for the pathogenicity of F. graminearum, and its deleted mutant could not cause disease in wheat (Son et al. 2011). However, the Δfpada1 mutant could infect wheat and the deficiency in pathogenicity might be due to reduction of growth.
In fungi, cell cycle regulation has been shown to be important in terms of growth and development, and this helps ensure that cells maintain their normal size, shape and nuclear number (Ahmadian et al. 2019; Jiang et al. 2016). The cyclin-dependent protein kinases CDKs are the central regulators of the eukaryotic cell cycle (Liu et al. 2015). The Cdc2 kinase in yeasts and filamentous fungi has a key regulatory role in the cell cycle. Unlike other fungi, F. graminearum have two Cdc2 genes, and the two Cdc2 orthologs play different roles in vegetative and infectious hyphae (Sudbery 2008). Cdc42 is a member of the Rho family of GTPases, which are required for hyphal growth in many fungi (Nozaki et al. 2018; Si et al. 2016). Here, we found that the FpAda1 deletion cells had abnormal nuclear morphology, and FpCdc2 and FpCdc42 were affected in expression.
In conclusion, our study demonstrated that the bZIP transcription factor plays important roles in growth, conidiation and pathogenesis in F. pseudograminearum. In addition, FpAda1 can affect cell cycle and the expression of FpCdc2 and FpCdc42.
References
Ahmadian M, Tyson JJ, Cao Y (2019) A stochastic model of size control in the budding yeast cell cycle. BMC Bioinform 20:322
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Alves MS, Dadalto SP, Goncalves AB, De Souza GB, Barros VA, Fietto LG (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int J Mol Sci 14:7815–7828
Amorim LLB, da Fonseca-Dos-Santos R, Neto JPB, Guida-Santos M, Crovella S, Benko-Iseppon AM (2017) Transcription factors involved in plant resistance to pathogens. Curr Protein Pept Sci 18:335–351
Aoki T, O’Donnell K (1999) Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognized as the Group 1 population of F. graminearum. Mycologia 91:597
Banerjee A, Roychoudhury A (2017) Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254:3–16
Bassilana M, Hopkins J, Arkowitz RA (2005) Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell 4:588–603
Bloom J, Cross FR (2007) Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol 8:149–160
Booher R, Beach D (1986) Site-specific mutagenesis of cdc2+, a cell cycle control gene of the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 6:3523–3530
Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N et al (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68:1–108
Boyce KJ, Hynes MJ, Andrianopoulos A (2001) The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J Bacteriol 183:3447–3457
Chen L, Geng X, Ma Y, Zhao J, Chen W, Xing X, Shi Y, Sun B, Li H (2019a) The ER lumenal Hsp70 protein FpLhs1 is important for conidiation and plant infection in Fusarium pseudograminearum. Front Microbiol 10:1401
Chen L, Tong Q, Zhang C, Ding K (2019b) The transcription factor FgCrz1A is essential for fungal development, virulence, deoxynivalenol biosynthesis and stress responses in Fusarium graminearum. Curr Genet 65:153
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103:10352–10357
Gardiner DM, Benfield AH, Stiller J, Stephen S, Aitken K, Liu C, Kazan K (2018) A high-resolution genetic map of the cereal crown rot pathogen Fusarium pseudograminearum provides a near-complete genome assembly. Mol Plant Pathol 19:217–226
Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H, Song W, Wang W, Wang Q, Lv R et al (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact MPMI 23:1053–1068
Guo M, Chen Y, Du Y, Dong Y, Guo W, Zhai S, Zhang H, Dong S, Zhang Z, Wang Y et al (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7:e1001302
He XJ, Fassler JS (2005) Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol Microbiol 58:1454–1467
Humphrey T, Pearce A (2005) Cell cycle molecules and mechanisms of the budding and fission yeasts. Methods Mol Biol 296:3–29
Hurst HC (1995) Transcription factors 1: bZIP proteins. Protein Profile 2:101–168
Jiang C, Xu JR, Liu H (2016) Distinct cell cycle regulation during saprophytic and pathogenic growth in fungal pathogens. Curr Genet 62:185–189
Kazan K, Gardiner DM (2018) Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: recent progress and future prospects. Mol Plant Pathol 19:1547–1562
Kong S, Park SY, Lee YH (2015) Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae. Environ Microbiol 17:1425–1443
Li H, Yuan H, Fu B, Xing X, Sun B, Tang W (2012) First report of Fusarium pseudograminearum causing crown rot of wheat in Henan, China. Plant Dis 96:1065
Li X, Wu Y, Liu Z, Zhang C (2017) The function and transcriptome analysis of a bZIP transcription factor CgAP1 in Colletotrichum gloeosporioides. Microbiol Res 197:39–48
Liu H, Zhang S, Ma J, Dai Y, Li C, Lyu X, Wang C, Xu JR (2015) Two Cdc2 kinase genes with distinct functions in vegetative and infectious hyphae in Fusarium graminearum. PLoS Pathog 11:e1004913
Lv W, Wu J, Xu Z, Dai H, Ma Z, Wang Z (2019) The putative histone-like transcription factor FgHltf1 is required for vegetative growth, sexual reproduction, and virulence in Fusarium graminearum. Curr Genet 65:981
Molina L, Kahmann R (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19:2293–2309
Moran KD, Kang H, Araujo AV, Zyla TR, Saito K, Tsygankov D, Lew DJ (2019) Cell-cycle control of cell polarity in yeast. J Cell Biol 218:171–189
Nozaki S, Furuya K, Niki H (2018) The Ras1-Cdc42 pathway is involved in hyphal development of Schizosaccharomyces japonicus. FEMS Yeast Res 18:31
Obanor F, Neate S, Simpfendorfer S, Sabburg R, Wilson P, Chakraborty S (2013) Fusarium graminearum and Fusarium pseudograminearum caused the 2010 head blight epidemics in Australia. Plant Pathol 62:79–91
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rincon SA, Estravis M, Perez P (2014) Cdc42 regulates polarized growth and cell integrity in fission yeast. Biochem Soc Trans 42:201–205
Sendinc E, Jambhekar A, Shi Y (2015) Remodeling your way out of cell cycle. Cell 162:237–238
Si H, Rittenour WR, Harris SD (2016) Roles of Aspergillus nidulans Cdc42/Rho GTPase regulators in hyphal morphogenesis and development. Mycologia 108:543–555
Son H, Seo YS, Min K, Park AR, Lee J, Jin JM, Lin Y, Cao P, Hong SY, Kim EK et al (2011) A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog 7:e1002310
Sudbery PE (2008) Regulation of polarised growth in fungi. Fungal Biol Rev 22:44–55
Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG (2008) A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci USA 105:2934–2939
Tian C, Li J, Glass NL (2011) Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157:747–759
Warren AJ (2002) Eukaryotic transcription factors. Curr Opin Struct Biol 12:107–114
Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T (2006) A systems approach to mapping DNA damage response pathways. Science 312:1054–1059
Zhou H, He X, Wang S, Ma Q, Sun B, Ding S, Chen L, Zhang M, Li H (2019) Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China. Environ Microbiol 21:2740–2754
Acknowledgements
This project is supported by grants from the National Key R&D Plan of China (2017YFD0301104), the Special Fund for Agro-Scientific Research in the Public Interest (201503112) and the Key Research Project of Colleges and Universities in Henan Province (16A210007).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2019_1042_MOESM1_ESM.jpg
Fig. S1 Western blotting of FpAda1 from transformed mycelia. Total protein was extracted from mycelia grown in vitro. FpAda1 was detected with anti-FLAG antibody. FpAda1 protein is ~ 80 kDa
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, L., Ma, Y., Zhao, J. et al. The bZIP transcription factor FpAda1 is essential for fungal growth and conidiation in Fusarium pseudograminearum. Curr Genet 66, 507–515 (2020). https://doi.org/10.1007/s00294-019-01042-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-019-01042-1