Abstract
Tra1 is a component of the Saccharomyces cerevisiae SAGA and NuA4 complexes and a member of the PIKK family, which contain a C-terminal phosphatidylinositol 3-kinase-like (PI3K) domain followed by a 35-residue FATC domain. Single residue changes of L3733A and F3744A, within the FATC domain, resulted in transcriptional changes and phenotypes that were similar but not identical to those caused by mutations in the PI3K domain or deletions of other SAGA or NuA4 components. The distinct nature of the FATC mutations was also apparent from the additive effect of tra1-L3733A with SAGA, NuA4, and tra1 PI3K domain mutations. Tra1-L3733A associates with SAGA and NuA4 components and with the Gal4 activation domain, to the same extent as wild-type Tra1; however, steady-state levels of Tra1-L3733A were reduced. We suggest that decreased stability of Tra1-L3733A accounts for the phenotypes since intragenic suppressors of tra1-L3733A restored Tra1 levels, and reducing wild-type Tra1 led to comparable growth defects. Also supporting a key role for the FATC domain in the structure/function of Tra1, addition of a C-terminal glycine residue resulted in decreased association with Spt7 and Esa1, and loss of cellular viability. These findings demonstrate the regulatory potential of mechanisms targeting the FATC domains of PIKK proteins.
Similar content being viewed by others
Introduction
Tra1 is a component of the yeast SAGA and NuA4 complexes, being the principal component that interacts with transcription activators (Bhaumik et al. 2004; Brown et al. 2001; Fishburn et al. 2005; Reeves and Hahn 2005). Tra1 is essential for viability in Saccharomyces cerevisiae (Saleh et al. 1998). Its mammalian homolog TRRAP is required for early embryonic development (Herceg et al. 2001) and the function of key cellular regulators such as c-Myc, p53, E2F1, β-catenin, and BRCA1 (reviewed by Murr et al. 2007). Tra1 and TRRAP are members of the phosphatidylinositol 3-kinase (PI3K) related kinase (PIKK) family, which also includes ATM, ATR, DNA-PKcs, TOR, and SMG-1. All of these molecules are important players in stress response, particularly related to DNA damage, cell growth, and proliferation (Abraham 2004). Tra1/TRRAP retains the PI3K domain, but the protein kinase activity demonstrated for many members of the family has not been found (McMahon et al. 1998; Saleh et al. 1998; Vassilev et al. 1998).
The SAGA complex is engaged in a number of nuclear processes. Its roles include facilitating recruitment of the transcriptional preinitiation complex (Bhaumik and Green 2001, 2002; Larschan and Winston 2005), promoting nucleosome eviction (Govind et al. 2007) and replication-coupled nucleosome assembly (Burgess et al. 2010). These regulatory functions occur through the acetylation of nucleosomal histones H2B, H3, and Htz1 by the component protein Gcn5 (Grant et al. 1997; Millar et al. 2006; Ruiz-Garcia et al. 1997; Suka et al. 2001), the deubiquitylation of histone H2B by Ubp8 (Henry et al. 2003), and interaction with the basal transcriptional machinery (Dudley et al. 1999; Mohibullah and Hahn 2008; Saleh et al. 1997). The presence of the nuclear pore component Sus1 within SAGA also links the complex with mRNA export (Kohler et al. 2006, 2008).
The catalytic subunit of the NuA4 complex, Esa1, is essential for viability in S. cerevisiae and acetylates histones H2A, H4, and Htz1 (Allard et al. 1999; Millar et al. 2006). Acetylation by Esa1 is required for transcriptional regulation (Allard et al. 1999) and the DNA-damage response (Bird et al. 2002; Choy and Kron 2002; Downs et al. 2004). A subset of the other NuA4 component proteins, Eaf2, Act3/Arp4, Act1, and Yaf9, are shared with the Swr1 complex that introduces Htz1 into chromatin (Bird et al. 2002; Choy and Kron 2002; Downs et al. 2004; Krogan et al. 2003, 2004).
We previously characterized a class of tra1 alleles having mutations within the PI3K domain (Mutiu et al. 2007a). The most severe allele, tra1-SRR3413 is a triple alanine scanning mutation that alters the serine-arginine-arginine residues found at positions 3413 to 3415. The changes in gene expression in the tra1-SRR3413 strain partially overlap those seen in strains with deletions of SAGA or NuA4 components and result in phenotypes consistent with the involvement of Tra1 in cell wall stability and stress response. Synthetic genetic array analysis identified genetic interactions of tra1-SRR3413 with genes involved in gene expression, mitochondrial function, and membrane sorting/protein trafficking (Hoke et al. 2008b). In addition, tra1-SRR3413 shows generation-dependent telomere shortening, a phenotype not seen with deletions of SAGA or NuA4 components (Mutiu et al. 2007a).
The extreme C-terminus of the PIKK proteins contains a 35-amino acid residue FATC domain (FRAP-ATM-TRRAP C-terminus; Bosotti et al. 2000). For ATM, DNA-PKcs, mTOR, and SMG-1, the FATC domain is necessary for the kinase activity of the adjacent PI3K domain (Beamish et al. 2000; Morita et al. 2007; Priestley et al. 1998; Sun et al. 2007; Takahashi et al. 2000). In addition, the FATC domain of ATM is required for interaction with Tip60, the mammalian homolog of Esa1 (Sun et al. 2005). ATR, TRRAP, and DNA-PKcs FATC domains can substitute for the native domain of ATM, restoring kinase activity and interaction with Tip60 (Jiang et al. 2006); however, functional equivalency across the family is not absolute since the ATM FATC domain cannot replace that of mTOR (Takahashi et al. 2000). A solution structure for the isolated FATC sequence of S. cerevisiae Tor1 consists of an α-helix with a C-terminal disulfide bonded loop (Dames et al. 2005). The generality of this structure is unclear given that the cysteine residues that form the disulfide bond are not present in other PIKK family members.
The goal of this study was to identify features of the FATC domain that are important for the function of the Tra1/TRRAP molecules. By analyzing mutations within the FATC domain of Tra1, we show that the FATC domain and precise positioning of the C-terminal carboxyl group are required for function. Addition of a C-terminal glycine resulted in loss of viability and altered association with NuA4 and SAGA components. Alanine substitutions at L3733 or F3744 resulted in growth phenotypes and transcriptional changes related, but not identical, to those within the PI3K domain. Tra1-L3733A was characterized in more detail, as it caused the most specific growth defects. We suggest that the functional changes of Tra1-L3733A are due to a role for the FATC domain in maintaining a stable form of the protein since the steady state level of Tra1-L3733A was 25% of that seen for wild-type Tra1, and suppressor mutations that partially restored function of Tra1-L3733A increased its concentration to a similar extent. These findings demonstrate the importance of the FATC domain in the structure/function of Tra1 and emphasize the pronounced consequences of any regulatory mechanism that targets the FATC domain of the Tra1/TRRAP proteins.
Materials and methods
Yeast strains and growth
Yeast strains are listed in Table 1. CY4060 is a derivative of BY4743 in which one copy of TRA1 has been gene replaced with tra1-L3733A that contains a HIS3 allele at the downstream BstBI site. CY4060 was sporulated to generate MATa and MATα haploid strains (CY4103 and CY4057, respectively) that were then crossed to deletion derivatives of BY4741 and BY4742 and sporulated to analyze the double mutant strains. CY4353, CY4318, CY4324, and CY4350 are similarly engineered strains containing wild-type TRA1, tra1-A3734S, tra1-F3740A, and tra1-F3744A.
TRA1 alleles contained on TRP1 centromeric plasmids were transformed into CY1021 (Saleh et al. 1998) and the wild-type copy on a URA3-centromeric plasmid displaced by plasmid shuffling. Growth comparisons were performed on plates at 30° unless stated otherwise. Assays were performed in duplicate on independently constructed strains. Scoring of FATC domain mutations was relative to CY2706, which contains TRA1 WT , the background allele used to construct the mutations. TRA1 WT is N-terminally myc9-tagged and contains a BamHI site that converts N3580A.
TAP-tagged ADA2 (YSC1178-7500046), SPT7 (YSC1178-7499287) and ESA1 (YSC1178-7502907) (Ghaemmaghami et al. 2003) BY4741, and BY7042 (yaf9::Kan r) were purchased from Open Biosystems. These strains were made trp1::URA3 and leu2::HIS3 using pTU10 and pLH7, respectively (Cross 1997).
DNA constructs
The PHO5-LacZ reporter constructs in the LEU2 centromeric plasmid YCp87 were described previously (Mutiu et al. 2007b). Rpl35a-LacZ was similarly constructed using the oligonucleotides indicated in Table 2. A HIS3-LacZ fusion regulated by two STRE elements (SRE/his3-LacZ) in place of the Gcn4 binding site was constructed by annealing oligonucleotides 5669-1 and 5669-2 and inserting the fragment into the EcoRI and SacI sites of his3-Δ88-LacZ (Brandl et al. 1993).
myc 9 -TRA1-YCplac111 was constructed through consecutive ligation of oligonucleotides 5088-1 and 5088-2 (see Table 2) into myc-TRA1-YCplac111 (Saleh et al. 1998). A BamHI site at position 10734 of TRA1 was introduced by two-step PCR using oligonucleotide pairs of 2583-1 with 2346 and 2583-2 with 2323-2 and cloned as an ApaI–SacI restriction fragment into full-length TRA1 to give myc 9 -TRA1 WT . Mutations of L3721D, D3722A, L3733A, D3737Y, and G3745 were similarly engineered. Initial reactions contained a listed oligonucleotide and the appropriate outside flanking primer with a unique cloning site (oligonucleotides 2346 and 4249-3). Fragments were moved into myc 9 -TRA1 WT -YCplac111 using BamHI–SacI restriction sites. T3714I and I3720D were serendipitously isolated in sequencing of random alleles.
To integrate tra1-A3727S, L3733A, F3740A, and F3744A into the genome, mutations were introduced into the 3′ SphI–FspI fragment of the gene and flanking region using the oligonucleotides listed in Table 2 and the terminal NcoI site for cloning. The DNA contained HIS3 at the BstBI site to allow selection in yeast. A plasmid copy of YHR100C was transformed into the strains to ensure that this gene was not affected by the integration.
A 495-bp fragment of the MET3 promoter flanked by PstI and NotI sites was cloned into the molecules expressing TRA1 after PCR with oligonucleotides 5295-1 and 5295-2. TAP-tagged TRA1 molecules were cloned into a LEU2 derivative of the YCpDed-TAP construct described previously (Mutiu et al. 2007a).
β-Galactosidase assays
Yeast strains containing RPL35a-LacZ were grown in YPD to an A 600 of ~1.5, pelleted, washed in LacZ buffer, and concentrated fivefold. β-Galactosidase was determined using ο-nitrophenol-β-d-galactosidase as substrate, standardizing to cell density (Ausubel et al. 1988). For analysis of PHO5-LacZ under inducing conditions, overnight cultures were washed three-times in water then grown 15 h in YPD depleted of phosphate (Han and Grunstein 1988). STRE/his3-LacZ and his3-Δ88-LacZ were assayed after growing tenfold dilutions of saturated cultures (from minimal media) for 15 h in YPD containing 4% ethanol.
RNA purification and gene profiling
Yeast cells, CY2706, and CY3003, were grown at 30° in YP media containing 2% glucose to an A 600 = 2.0. RNA was purified from 108 cells after glass bead disruption as described previously (Mutiu and Brandl 2005). RNA integrity numbers of greater than 8.9 were determined for each RNA sample using an Agilent 2100 Bioanalyzer at the London Regional Genomics Centre. mRNA-Seq libraries were constructed and sequencing were performed on the Illumina/Solexa Genome Analyzer II platform at the DNA Facility at Iowa State University. The CY2706 and CY3003 samples were each run on a single Illumina GAII lane, producing 13014880 reads for CY2706 and 11156078 reads for CY3003 of 35 nucleotides. The S. cerevisiae genome sequence and the general features format file (saccharomyces_cerevisiae.gff) were obtained from Saccharomyces Genome Database on May 1, 2009. The sequencing reads were mapped onto the genomic sequence using the novoindex and novoalign programs with the default parameters, except that reads mapping to two or more places in the genome were placed at one position at random. With this option novoalign marks a read as uniquely or repetitively mapping in the genome; only uniquely mapping reads were used for the subsequent analysis (83 and 84% for CY2706 and CY3003, respectively). Mapped reads were placed into bins composed of protein-coding genes, tRNA and rRNA genes as defined by the gff file. Only reads that did not overlap the start or end position of the gene were counted and reads mapping to the top and bottom strands were tabulated separately. The relative occurrence of each ORF annotated in the Saccharomyces Genome Database as a ratio of its length was calculated after normalization to 10 million reads for each sample, similar to the normalization outlined in Mortazavi et al. (2008). Genes with ≥0.05 reads per base pair of gene length were considered for further analysis. Agglomerative hierarchical clustering based on the average linkage of uncentered correlations was performed using CLUSTER 3.0 software (Eisen et al. 1998) on the profiles from strains within the compendium data set (Hughes et al. 2000) the data sets of strains containing deletions of NuA4 (Krogan et al. 2004) and SAGA components (Ingvarsdottir et al. 2005) and with tra1-SRR3413 (Mutiu et al. 2007a). Genes not appearing in at least two of the profiles were excluded. The data were visualized using MAPLETREE (http://rana.lbl.gov/EisenSoftware.htm).
RNA dot blots with probes for ADE17 and RPL4a/b (Table 2) were performed on Hybond-N membrane (Amersham) using 10 and 2.5 μg of total RNA, essentially as described by the manufacturer. Hybridizations were performed in buffer containing 5× standard saline citrate (SSC), 5× Denhardt’s solution, 0.5% sodium dodecyl sulphate (SDS), and 90 μg/ml denatured herring sperm DNA at 52°. Washes in 2× SSC plus 0.1% SDS and 1× SSC plus 0.1% SDS were performed at 42°.
Chromatin immunoprecipitation assays
Assays for acetylated histones were performed essentially as described previously (Hoke et al. 2008a). Cells were grown in YPD media to an A 600 ~ 2.0. Antibodies were purchased from Abcam Inc. (anti-H3, ab1791; anti-AcH4/K8, ab1760).
Western blotting
Yeast extract prepared by grinding in liquid nitrogen or by lysis with glass beads (Saleh et al. 1997) was separated by SDS-PAGE and transferred to PVDF membrane (Roche Applied Science). Anti-myc (Evan et al. 1985), anti-Mcm2 (Santa Cruz Biotechnology, Cat. # sc-6680; kindly supplied by Megan Davey), and anti-calmodulin-binding protein (CBP) antibodies (Millipore Corp., Cat. # 07-482) were used at ratios of 1:5000, 1:4000, and 1:1000, respectively. Secondary antibody (anti-Mouse IgG HRP, Promega; anti-Goat IgG HRP, Sigma; anti-Rabbit IgG HRP, Promega) used at a ratio of 1:10000 was detected using SuperSignal West Pico Chemiluminiscent Substrate (Thermo Scientific). Densitometric scanning of films was performed using AlphaImager 3400 software (Alpha Innotech, Inc.).
TAP purification
Whole cell extracts were prepared by grinding in liquid nitrogen (Saleh et al. 1997). Tandem affinity purification (Rigaut et al. 1999) with 1 l of extract grown in minimal media lacking tryptophan to an A 600 ~ 2 was carried out as described previously (Mutiu et al. 2007a).
Genome-wide localization studies
Genome-wide localization studies were performed essentially as described (Yu et al. 2004) for yeast strain CY2706 grown at 30° in YPD. Immunoprecipitations were performed in triplicate with 10 μl of anti-myc antibody (9E11) and using pan-mouse IgG Dynal beads (Invitrogen). Antibody was pre-incubated with the beads in 1× phosphate-buffered saline containing 5 mg/ml BSA for a minimum of 2 h. P values were calculated using an error model provided by Rosetta Resolver. The genome-wide occupancy was expressed as the ratio of fluorophore intensities from chromatin fragments enriched by immunoprecipitation versus that of the input chromatin fragments. Spots with a P value threshold of 0.02 and a ratio of intensity >1.0 were included in the final dataset (Online Resource 1).
Isolation of intragenic suppressors of tra1-L3733A
C-terminal sequences of tra1-L3733A downstream of the ApaI site at base pair 9175 were mutagenized by PCR, cloned back into the full-length molecule, and shuffled into yeast strain CY4018 by selection on 5-FOA. Individual colonies were selected for growth on YPD plates containing 4% ethanol; the plasmids were isolated, sequenced, and verified for plasmid dependency of the selection by repeating the selection process after transformation into CY1021.
Gal4 affinity chromatography
Interaction of myc9-Tra1 constructs from yeast strain CY2998 with recombinant activation domain of Gal4 was performed as described by Mutiu et al. (2007a).
Results
Characterization of mutations within the FATC domain of Tra1
The C-terminal region of the Tra1/TRRAP family contains three conserved domains: FAT (FRAP-ATM-TRRAP), phosphatidylinositol 3-kinase-like (PI3K) and FATC (FAT C-terminal; Fig. 1a). To identify key residues required for function, we introduced mutations into the FATC domain. To identify residues to target, we analyzed an alignment of the FATC domains of Tra1/TRRAP from five species (Fig. 1b, upper alignment) and an alignment of S. cerevisiae Tra1 with the FATC domains from members of the PIKK family (lower alignment). L3733 and A3727 (numbering for S. cerevisiae Tra1) are highly conserved throughout the PIKK family, as are hydrophobic residues at positions equivalent to I3720, I3724, F3740, W3743, and F3744. An acidic residue is conserved at D3737 within Tra1/TRRAP, but is aromatic in the broader family. Other positions are conserved in the fungal forms of Tra1. We constructed alleles of S. cerevisiae TRA1 with changes to these different classes of residues (see Fig. 1b). The L3733A change was of particular interest because the comparable mutation in SMG-1 results in loss of kinase activity (Morita et al. 2007). Some of the changes, for example, L3721D and D3737Y, were made to resemble the residues found in the PIKK family. Another allele, which we have termed tra1-G3745, was constructed with a glycine codon following the terminal phenylalanine codon to analyze the importance of the positioning of the terminal carboxyl group.
FATC domain mutants. a Schematic diagram of the architecture of the Tra1/TRRAP family indicating the approximate positions of the HEAT, FAT, PI3 K, and FATC domains (Bosotti et al. 2000; Murr et al. 2007). Numbering is for the 3744 residues of S. cerevisiae Tra1. b Sequence alignment of FATC domains from PIKK family proteins. Top panel Sequences are Saccharomyces cerevisiae Tra1 (NP_011967), Schizosaccharomyces pombe Tra2 (Q10064), Schizosaccharomyces pombe Tra1 (NP_595777), Neurospora crassa Tra1 (CAC18279), Drosophila melanogaster Nipped-A (NP_001097192), Homo sapiens TRRAP (NP_003487). Lower panel Sequences from Saccharomyces cerevisiae Tra1 (NP_011967), Tor1 (CAA52849) and Homo sapiens TRRAP (NP_003487), ATM (Q13315), ATR (Q13535), DNA-PKcs (P78527) and SMG-1 (NP_055907). Residues indicated between the alignments in bold capitals are those changes that were analyzed through plasmid shuffling. Lower case letters indicate residues analyzed by gene integration, see c. Numbering is for S. cerevisiae Tra1. Alignments were performed using the default parameters of the ClustalW utility [http://www.ebi.ac.uk/clustalw/] and the terminal 49 residues of each protein. c Growth of the tra1-L3733A strain. Yeast strains containing centromeric plasmids expressing tra1-L3733A and TRA1 WT were grown to saturation and ten-fold serial dilutions plated onto YP media containing 2% glucose and grown at 30° or 16°, or at 30° on YPD containing 5 μg/ml Calcofluor white (CW), 1 nM rapamycin, 20 μg/ml geneticin, or 4% ethanol. d Analysis of integrated tra1 alleles. TRA1 WT , tra1-L3733A, tra1-A2727S, tra1-F3740A and tra1-F3744A were integrated into BY4743. Haploids containing the integrated allele were obtained after sporulation. Saturated cultures were serially diluted and plated onto YPD at 30° or 37°, or at 30° on YPD containing 5 μg/ml Calcofluor white (CW), 1 nM rapamycin, or 6% ethanol. We note that the BY4742/4741 background is less sensitive to ethanol than KY320, so ethanol sensitivity was assayed at a concentration of 6%
The initial tra1 alleles analyzed (T3714I, I3730D, L3721D, D3722A, L3733A, D3737Y and G3745) were introduced on TRP1-centromeric plasmids into S. cerevisiae strain CY1021, which contains a disruption of the genomic copy of TRA1, complemented by wild-type TRA1 expressed from a URA3-containing centromeric plasmid. Interestingly, tra1-G3745 and to a lesser extent tra1-L3733A, resulted in slow growth in combination with the wild-type allele (not shown). The alleles were examined for their ability to support viability by shuffling out wild-type TRA1 on media containing 5-fluoroorotic acid. The six alleles with single residue changes supported growth, whereas tra1-G3745 did not. Of the viable strains, only the strain containing tra1-L3733A had obvious growth defects.
The tra1-L3733A allele resulted in several phenotypes shared with strains having mutations in the ada genes (Fig. 1c; Table 3). These phenotypes included slow growth on media containing ethanol, Calcofluor white, or tunicamycin. Interestingly, however, it did not display the classic ada phenotype of resistance to overexpression of VP16 (Berger et al. 1992). The tra1-L3733A allele decreased growth on media containing tert-butylhydroperoxide, or lacking inositol, both characteristics of defects in Spt function (Gansheroff et al. 1995). A dichotomy was seen for NuA4-related phenotypes, as the tra1-L3733A strain was sensitive to benomyl but not methylmethane sulphonate.
The phenotypes of the tra1-L3733A strain were similar but not identical to those of the tra1-SRR3413 strain (Mutiu et al. 2007a). Similarities included slow-growth on media containing ethanol, Calcofluor white, benomyl, rapamycin, geneticin, and chloramphenicol; whereas, differences in sensitivity to tert-butylhydroperoxide and tunicamycin were observed. In addition, defects in telomere maintenance or elongation were not observed using the plasmid linearization assay of Lundblad and Szostak (1989; not shown).
To examine the importance of the highly conserved alanine at 3727 and the hydrophobic residues, F3740 and F3744, we constructed yeast strains in which tra1-A3727S, tra1-F3740A, tra1-F3744A, as well as tra1-L3733A were integrated into the genome of the wild-type strain BY4741/4742 and analyzed growth under a variety of conditions (Fig. 1d). Similar to the plasmid copy, the integrated allele of tra1-L3733A, expressed from its native promoter, resulted in slow growth at 37° and in media containing Calcofluor white, rapamycin, and ethanol. The tra1-A3727S strain was slightly sensitive to each of these conditions; in comparison, the tra1-F3740A strain was relatively unaffected. Mutation of the terminal phenylalanine to alanine (F3744A) resulted in a general reduction in viability in all conditions assayed, including rich media at 30°. This reduced viability demonstrates the importance of the terminal residue for Tra1 function and is consistent with the inability of tra1-G3745 to support viability. Interestingly, tra1-F3744A did not result in as pronounced specific phenotypes as seen with tra1-L3733A. In fact, the tra1-F3744A strain was slightly less sensitive to Calcofluor white and ethanol than the tra1-L3733A strain.
Transcriptional effects of mutations within the FATC domain
The effects of the FATC domain mutations on transcription were initially assayed by determining the expression of the SAGA and NuA4-dependent PHO5 promoter (Barbaric et al. 2003; Nourani et al. 2004). LacZ assays were performed under inducing conditions with the integrated tra1 alleles (wild-type, L3733A, A3727S, F3740A and F3744A; Fig. 2a). The effect of these alleles on PHO5 expression followed a similar pattern to their effects on growth. Tra1-L3733A and F3744A reduced PHO5 expression to ~20% of wild-type. Tra1-A3727S and F3740A had a more modest effect, reducing expression to ~60% of wild-type. We note that this compares with PHO5-LacZ expression of <5% of the wild-type level seen upon deletion of either the SAGA component, Spt7 or NuA4 component, Yng2 (not shown).
Gene expression in the tra1-L3733A strain. a The promoter region of PHO5 was cloned as a LacZ reporter fusion into the LEU2 centromeric plasmid YCp87 and transformed into yeast strains CY4353 (TRA1), CY4318 (tra1-A3727S), CY4103 (tra1-L3733A), CY4324 (tra1-F3740A) and CY4350 (tra1-F3744A), containing a plasmid copy of YHR100C. β-Galactosidase activity was determined after growth in low phosphate media for 15 h at 30°. Expression is shown as a percentage of that found for CY4353. Measurements were made in triplicate with the standard deviation indicated. b Expression from stress response elements. A cassette of two STRE elements was cloned into the EcoRI and SacI sites of his3-Δ88-LacZ (Brandl et al. 1993) to give STRE-his3-LacZ. These elements replace the normal Gcn4 binding site in HIS3. STRE-his3-LacZ and his3-Δ88 were transformed into CY2706 and CY3003, and β-galactosidase assays performed after growth of cells in YPD containing 4% ethanol. c Expression levels determined by gene profiling (GP, mRNA-Seq) as compared with RNA dot blots (ADE17 and RPL4a/b) and LacZ reporter fusions (PHO5 and RPL35a). RNA was prepared from yeast strains CY3003 (tra1-L3733A) and CY2706 (TRA1 WT ). mRNA-Seq libraries were constructed and sequencing performed on the Illumina/Solexa Genome Analyzer II platform. Comparisons between the strains were made as outlined in “Materials and methods”. Similarly prepared RNA was spotted onto Hybond-N membrane and probed with single-stranded DNAs complementary to ADE17 and RPL4a/b RNA. Hybridization was detected by autoradiography and quantitated using AlphaImager 3400 software and shown as a percentage of wild-type expression. Expression of PHO5-LacZ and RPL35a-LacZ fusion reporters were determined after growth of CY3003 and CY2706 in YPD. Assays were performed in triplicate
The phenotypes of the tra1-L3733A strain suggested a partial inability to respond to environmental change and stress. Though multiple factors are involved in the cellular response to stress, a general stress response involves transcriptional induction upon binding of transcription factors Msn2 and Msn4 to stress response elements (STRE elements) (Gasch et al. 2000; Harbison et al. 2004; Martinez-Pastor et al. 1996). To determine if tra1-L3733A affected activation through STRE elements, we constructed a hybrid promoter containing two STRE elements at the position of the Gcn4-binding site in the HIS3 promoter, and assayed transcription when cells were grown in YPD containing 4% ethanol. As shown in Fig. 2b, expression of STRE/his3 was reduced threefold in the tra1-L3733A strain. The effect of the L3733A mutation was dependent on the stress response elements as the comparable promoter lacking the STRE elements (his3-Δ88) was only slightly affected.
To analyze for transcriptional effects of the tra1-L3733A allele on a broader scale, we compared the gene expression profiles of wild-type (CY2706) and tra1-L3733A (CY3003) strains grown in YPD using next generation sequencing. The full data set has been submitted to the Gene Expression Omnibus at the National Center for Biotechnology: accession number GSE18591. After normalization, expression of 11 genes was elevated ≥2-fold; 79 genes were decreased ≥2-fold (Table 4). While no over-riding patterns were apparent, of the 11 genes with elevated expression, HSP26, PIR3, DDR2, and GRE1 have roles in the cellular response to stress. Confirmation of the general profile seen by sequencing was obtained by the analysis of LacZ-reporter fusions and dot blotting for selected genes as shown in Fig. 2c.
To address whether the expression changes determined for tra1-L3733A resembled patterns seen with other mutations, we performed a hierarchical cluster analysis with the compendium dataset of Hughes et al. (2000), the datasets of strains containing deletions of NuA4 (Krogan et al. 2004) and SAGA components (Ingvarsdottir et al. 2005), and with the dataset of the PI3K-domain mutation tra1-SRR3413 (Mutiu et al. 2007a) (Fig. 3a). Of the approximately 300 comparisons in the analysis, the gene expression profile of the strain containing tra1-L3733A clustered closest to tra1-SRR3413. Other components of SAGA and NuA4 did not cluster within this leaf suggesting that the effects of tra1-L3733A result from the combined alteration of SAGA and NuA4 complexes and/or that Tra1 has one or more roles outside these complexes.
Gene expression in the tra1-L3733A strain. a Hierarchical cluster analysis. Comparisons were initially made with the compendium data set (Hughes et al. 2000), profiles from strains with deletions of NuA4 (Krogan et al. 2004) and SAGA components (Ingvarsdottir et al. 2005), and with tra1-SRR3413 (Mutiu et al. 2007a). The diagram shows those profiles clustering in closest proximity to tra1-L3733A. Gene families are indicated on the right. b Correlation between degree of Tra1-binding and average transcriptional frequency. Genome-wide localization studies were performed in triplicate for yeast strain CY2706 (myc 9 -TRA1) grown at 30° in YPD, essentially as described (Yu et al. 2004). The average transcription frequency of each gene (Holstege et al. 1998) is plotted versus the relative binding of myc 9 -TRA1
The genome-wide occupancy profile of myc9-Tra1 was determined to help assess whether the effect of Tra1 on gene expression is direct. As shown in Fig. 3b, there is a positive correlation between genomic occupancy of Tra1 and transcriptional frequency (Holstege et al. 1998), suggesting that Tra1 is recruited to actively transcribed genes. In addition, there was a positive correlation for the top quartile of Tra1 binding and Fhl1 (p = 2.2 × 10−7) and Rap1 (p = 1.1 × 10−5), likely related to the involvement of these factors and the NuA4 complex in regulating expression of ribosomal protein genes (Lieb et al. 2001; Rudra et al. 2005; Reid et al. 2000).
Intragenic suppressors of tra1-L3733A
As a tool to evaluate mechanisms by which the L3733A mutation may affect Tra1 function, we selected random intragenic suppressor mutations that enable growth on media containing ethanol. A library of approximately 200 independent alleles was constructed by PCR mutagenesis of the C-terminal 2060 base pairs of the tra1-L3733A allele. Mutations N3677D and T3716A were able to partially suppress the slow growth in YPD containing 4% ethanol caused by the L3733A mutation (Fig. 4a) and restore transcription of PHO5-LacZ to approximately 70 and 80% of wild type, respectively (Fig. 4b). Both of the suppressor mutations occur at positions that are not highly conserved within the Tra1/TRRAP family. N3677 is at the C-terminal end of the PI3 K domain, while T3716 is within the FATC domain.
Intragenic supressors of the L3733A mutation. a Yeast strains CY3003 (tra1-L3733A), CY2706 (TRA1 WT ), CY4055 (tra1-L3733A/N3677D) and CY4056 (tra1-L3733A/T3716A) were grown in minimal media and tenfold serial dilutions spotted onto a YPD plate containing 4% ethanol. b PHO5-LacZ expression. The above strains containing YCp87-PHO5-LacZ were grown to saturation in minimal media lacking leucine, diluted sixfold in YPD depleted of phosphate and grown for 15 h at 30°. β-Galactosidase activity was calculated for each strain in triplicate and plotted as a percentage of that found for CY2706
Expression of Tra1-L3733A and Tra1-G3745
We used Western blotting to determine the steady-state levels of N-terminally myc9-tagged Tra1 in crude extracts of yeast strains CY2706 (Tra1WT expressed from the DED1 promoter) and CY3003 (Tra1-L3733A expressed from the DED1 promoter). As shown in Fig. 5a, Tra1-L3733A was reduced compared with wild-type Tra1. The profile of proteolytic products also differed for the wild-type and mutant proteins (compare lanes 2 and 4). A similar reduction of Tra1-L3733A was seen when cells were disrupted under denaturing conditions (not shown). As shown in Fig. 5b and quantified in Fig. 5c, the second site mutations N3677D and T3716A partially restored Tra1 levels and to an extent that paralleled their restoration of function. This correlation suggests that the phenotype of tra-L3733A is related to the reduced steady-state level of the protein.
Expression of the tra1-L3733A and tra1-G3745 alleles. a Whole cell extracts were prepared from yeast strain KY320 (−ve, lane 1) or strains expressing myc9-tagged versions of Tra1WT (lanes 2, 3) or Tra1-L3733A (lanes 4, 5). Extract was separated by SDS PAGE (5%) and Western blotted with anti-myc antibody. The upper panel is the Western blot with anti-myc antibody. The full length myc9-Tra1of ~450 kDa and the mobility of a 250-kDa marker protein are indicated with an arrow and dot, respectively. The lower panel is a portion of an equivalent gel stained with Coomassie Brilliant Blue (CBB). Lanes 1, 3, and 5 contain 50 μg of protein; lanes 2 and 4, 100 μg. b Levels of Tra1-L3733A suppressors. CY2706 (myc9-Tra1WT, lanes 1, 2), CY3003 (myc9-Tra1-L3733A, lanes 3, 4), CY4055 (myc9-Tra1-N3677D/L3733A lanes 5, 6), and CY4056 (myc9-Tra1-T3716A/L3733A lanes 7, 8) were grown to A 600 = 2.0. Crude protein was isolated by bead lysis and 50 μg (even numbered lanes) and 100 μg (odd numbered lanes) was separated by SDS-PAGE and Western blotted with anti-myc or anti Mcm2 antibodies. Expression of tra1-G3745. c Quantitation of Tra1 levels. The Western blot as in B was performed in triplicate with independently grown cultures. Relative amounts of Tra1 were determined by densitometry using AlphaImager 3400 software. d Histone H4 acetylation. Chromatin was isolated from yeast strains CY2706 (TRA1 WT , lower panel) or CY3003 (tra1-L3733A, upper panel) grown to A 600 = 2.0 in YPD. Chromatin was immunoprecipitated with antibody to histone H3 (lanes 2–4) and histone H4 acetylated at K8 (lanes 5–7), then analyzed by PCR with primers for PHO5 promoter sequences. Consecutive samples represent twofold serial dilutions of the DNA to enable quantitation. Lane 1 is a mock experiment with no histone antibody performed with a wild-type extract at 2× concentration
The cellular concentration of myc9-Tra1-G3745 was analyzed in a strain containing untagged wild-type Tra1 since tra1-G3745 does not support viability. The extreme slow growth of this strain made recovery of the protein difficult. We estimate that Tra1-G3745 was present at a level approximately 5% of wild-type (not shown) suggesting that the precise location of the C-terminal carboxyl group is critical for the stability of Tra1.
The NuA4 complex is localized to the PHO5 promoter prior to gene activation (Nourani et al. 2004). As another measure of the relative level of Tra1-L3733A in vivo, we determined the extent of histone H4 acetylation at the PHO5 promoter after growth of TRA1 WT and tra1-L3733A strains in YPD. Chromatin immunoprecipitations were performed with anti-acetylated histone H4/K8 antibody and to allow normalization, with anti-histone H3 antibody. As shown in Fig. 5d, under conditions in which total histone H3 was relatively unchanged (lanes 2–4), the L3733A mutation reduced histone H4 acetylation at PHO5 by approximately threefold (lanes 5–7).
If the effects of the L3733A mutation result primarily from decreased stability of Tra1, we would expect that reducing the wild-type protein would cause a similar phenotype. Cells containing wild-type Tra1 under control of the methionine-repressed MET3 promoter (Mao et al. 2002) (MET3-Tra1WT) were grown in minimal media with increasing concentrations of methionine and in the presence or absence of 3% ethanol. As shown in Fig. 6, in media lacking methionine MET3-Tra1WT supported growth in both media at a level comparable to DED1-expressed Tra1WT. In as little as 5 μM methionine, reduced expression of MET3-Tra1WT resulted in decreased growth of the strain in YPD and increased sensitivity to ethanol, which resembled that seen for the L3733A mutation (expressed from the DED1 promoter). We note that at elevated concentrations of methionine, fast-growing suppressors were evident with the strain containing MET3-Tra1WT, likely arising from derepression of the MET3 promoter or increased plasmid copy number. Given the number of generations required to obtain detectable amounts of Tra1, these suppressors made it difficult to compare the exact level of Tra1 in the presence of methionine.
Reduced expression of Tra1 results in ethanol sensitivity. Yeast strains CY3003 (DED1 promoter: tra1-L3733A), CY2706 (DED1 promoter: TRA1 WT ), and CY3021 (MET3 promoter: TRA1 WT ) were grown in minimal media lacking methionine, and then serial dilutions were spotted onto minimal plates containing the indicated concentration of methionine and for the lower panel 3% ethanol
Molecular interactions of Tra1-L3733A and Tra1-G3745
The ability of Tra1-L3733A to associate with SAGA and NuA4 components was compared to wild-type Tra1 and the phenotypically neutral Tra1-L3721D. myc9-tagged Tra1-L3721D, Tra1-L3733A, and Tra1WT were expressed in a strain containing TAP-ADA2 (Ghaemmaghami et al. 2003), and tandem affinity purification performed on crude yeast extracts. As shown in Fig. 7a, Tra1-L3721D and Tra1-L3733A co-purified with Ada2 at levels comparably to wild-type Tra1. Similarly, neither mutation affected interaction with TAP-Spt7 or with TAP-Esa1 (Fig. 7b). To determine if the L3733A mutation affects Tra1’s ability to interact with transcriptional activators, we analyzed the binding of Tra1-L3733A to the activation domain of Gal4 (Gal4AD). Myc9-tagged Tra1WT and Tra1-L3733A were purified via association with TAP-tagged Ada2. The affinity-purified SAGA complex was then chromatographed on GST-Gal4AD columns and the association of Tra1 determined after elution with glutathione by Western blotting. As shown in Fig. 7c, Tra1-L3733A interacted with the activation domain of Gal4 to approximately the same extent as wild-type Tra1.
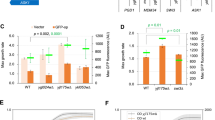
Protein interactions of Tra1-L3733A and Tra1-G3745. a Interaction with Ada2. Crude extracts were prepared from an ADA2-TAP strain (Ghaemmaghami et al. 2003) expressing myc9-Tra1WT (lane 1), myc9-Tra1-L3721D (lane 2), myc9-Tra1-L3733A (lane 3) or only the genomic TRA1 (−ve, lane 4). Volumes corresponding to equal amounts of myc-tagged Tra1 were subject to tandem affinity purification and equal volumes Western blotted using anti-myc antibody (upper panel) or anti-CBP (lower panel). b Interaction with Spt7 and Esa1. Crude extracts of SPT7-TAP and ESA1-TAP strains expressing myc9-Tra1WT, myc9-Tra1-L3721D, or myc9-Tra1-L3733A were analyzed as above and scanned using AlphaImager 3400 software. The ratio of the Spt7 and Esa1-associated to input Tra1 is presented as percent of wild type. The experiments were performed in triplicate with the associated standard error indicated. c Interaction of Tra1-L3733A with GST-Gal4AD. myc9-Tra1WT or myc9-Tra1-L3733A was expressed in yeast strain CY2998 containing TAP-tagged Ada2. After CBP affinity purification approximately equal amounts of Tra1WT (lanes 1, 2) or Tra1-L3733A (lanes 3, 4), as determined by Western blotting, were incubated with GST-Gal4AD bound to glutathione-Sepharose. After washing, three fractions were eluted with glutathione and Western blotted for the presence of Tra1WT (lanes 5, 7 and 9) or Tra1-L3733A (lanes 6, 8 and 10). The levels of bound Tra1WT and Tra1-L3733A correlated with the elution profile of GST-Gal4AD (not shown). d Tra1- G3745. Crude extracts were prepared from the ESA1-TAP, SPT7-TAP, or ADA2-TAP strains (Ghaemmaghami et al. 2003) expressing myc 9 -TRA1 WT or myc 9 -TRA1-G3745. Extracts were tandem affinity purified. Equal volumes were separated by SDS PAGE and Western blotted using anti-myc antibody to detect Tra1 or anti-CBP for Esa1, Spt7, or Ada2. AlphaImager 3400 software was used for densitometric analysis. The ratio between the purified and the input amounts is presented as percent of wild type. The experiments were performed in duplicate with the range as indicated
Since tra1-G3745 does not support viability, interaction of myc9-Tra1-G3745 with TAP-tagged Esa1, Spt7, and Ada2, was analyzed in strains also containing untagged wild-type Tra1. Crude extracts were tandem affinity purified and the level of myc9-Tra1 determined by Western blotting. After normalizing for the level of input Tra1, the amount of Tra1-G3745 co-purifying with Spt7 and Esa1 was diminished to <5%, of that found for wild-type Tra1 (Fig. 7d). Normal positioning of Tra1’s C-terminal carboxyl group is thus required for formation of SAGA and NuA4 complexes. Interestingly, the additional C-terminal glycine only partially reduced (~70%) the interaction with Ada2, suggesting that Tra1 has more than one interaction site with components of SAGA.
Genetic interactions of tra1-L3733A
To investigate the relationship between the PI3K and FATC domains, we constructed the double-mutant allele, tra1-SRR3413/L3733A and examined its ability to support viability after plasmid shuffling in yeast strain CY1021. Transformation of a TRP1 centromeric plasmid expressing Tra1-SRR3413/L3733A resulted in slow-growing colonies, suggesting a dominant negative effect of this allele (not shown). In addition, no colonies possessed the double-mutant allele (tra1-SRR3413/L3733A) after plasmid shuffling on 5-FOA. We conclude that the tra1-SRR3413/L3733A allele does not support viability and that the effects of PI3K and FATC domain mutations are additive.
tra1-L3733A was introduced into a group of the knock-out collection of strains (Winzeler and Davis 1997) to examine genetic interactions with SAGA and NuA4 component genes (Table 5). Growth of the double mutants was compared with either single mutant on YPD and YPD containing 3% ethanol. (As shown above, in the BY4741/4742 strain background tra1-L3733A alone only causes a minor growth defect in 3% ethanol; sensitivity is seen at 6% ethanol.) In YPD media, synthetic slow growth/lethality was observed with deletions of some but not all components. tra1-L3733A was synthetically lethal with spt20Δ and severe slow growth was seen with ada1Δ tra1-L3733A and eaf1Δ tra1-L3733A. Ada1 and Spt20 have roles in the structural integrity of SAGA (Sterner et al. 1999; Wu and Winston 2002), whereas Eaf1 is required for the integrity of the NuA4 complex (Auger et al. 2008; Babiarz et al. 2006). Similar synthetic lethality is found for double mutants of ada1Δ and spt20Δ with deletions of NuA4 components, and eaf1Δ with deletions of SAGA components (Lin et al. 2008, Mitchell et al. 2008). tra1-L3733A also resulted in synthetic slow growth in combination with disruptions of gcn5, ada2, ngg1, sgf29, and sgf73. In contrast, additive growth defects on YPD were not as pronounced with double mutants of tra1-L3733A with spt3Δ, spt8Δ, sgf11Δ, ubp8Δ, eaf3Δ, and eaf7Δ. Synthetic slow growth was observed for all the deletion combinations when cells were grown in media containing 3% ethanol. This implies that fully functional NuA4 and SAGA complexes are required under conditions of stress and that the effects of tra1-L3733A are additive with all functions of these complexes.
Larschan and Winston (2001) found that deletions of hda1 and nhp10 suppress phenotypes resulting from disruption of spt20, an integral component of the SAGA complex. To determine if the effect of tra1-L37733A is related to a similar loss of function as spt20, we analyzed whether hda1Δ0 and nhp1Δ0 suppress tra1-L3733A. Growth of double-mutant strains was analyzed on YPD at 16°, 30°, and 37° and YPD containing 6% ethanol (Fig. 8). Under none of these conditions was the slow growth caused by tra1-L3733A suppressed by deletion of either hda1 or nhp10. This result supports the view that the phenotypes arising from tra1-L3733A are not due solely to Tra1’s action in the SAGA complex.
Deletion of hda1 or nhp10 does not suppress tra1-L3733A. Strains deleted for hda1 or nhp10 were generated by sporulation of diploid deletion strains obtained from Open Biosystems. Double mutants with tra1-L3733A were obtained by mating with CY4057 or CY4103 and sporulation. The wild-type strain BY4742, and the single- and double-mutant strains were grown in YPD and serial dilutions spotted onto YPD at 30°, 16° or 37°, or YPD containing 6% ethanol at 30°
Discussion
Tra1 functions revealed by FATC domain mutations
Our studies demonstrate the consequences of reduced Tra1 function on gene expression. Mutation of L3733A resulted in decreased activation of PHO5 and STRE/his3 promoters, and a twofold or greater change in expression of ~90 genes in rich media. The effects of tra1-L3733A are likely mediated through partial loss of both SAGA and NuA4 function rather than loss of either individual complex. This is consistent with the phenotypic similarities between the tra1-L3733A strain and strains with deletions of components of NuA4 and SAGA, the additive effects of these mutations, and the inability of the tra1-L3733A allele to be suppressed by deletion of either hda1 or nhp10.
When considered together, the phenotypes displayed by the tra1-FATC mutant strains reveal a role for Tra1 in responding to a variety of stress conditions. Growth defects included temperature sensitivity and slow growth in media containing ethanol, calcofluor white, tert-butylhydroperoxide, and tunicamycin. The latter three indicate deficiencies in pathways required for cell wall integrity, response to oxidative stress and the unfolded protein response, respectively. A requirement for Tra1 in responding to nutrient levels is apparent from the sensitivity to rapamycin. As evident by the reduced expression from PHO5 and STRE/his3 promoters in the tra1-L3733A strain, the stress-related phenotypes may be due to the inability of the FATC mutants to activate the expression of genes required to manage the stress. This interpretation agrees with the general finding that many SAGA-regulated genes fall into the category of stress-induced (Huisinga and Pugh 2004).
The tra1-L3733A strain did not possess all the phenotype characteristic of deletions of SAGA and NuA4 components. This suggests that some activities of the SAGA and NuA4 complexes are relatively unaffected by reduced levels of Tra1. For example unlike strains with deletions of the ADA genes (Berger et al. 1992), the tra1-L3733A strain is sensitive to VP16 overexpression. This could occur if sensitivity to VP16 requires minimal SAGA-mediated acetylation, or alternatively, sensitivity can result from Gcn5 activity independent of Tra1. The possibility of the latter is consistent with biochemical evidence for an Ada complex (Eberharter et al. 1999; Saleh et al. 1997).
The FATC domain is required for Tra1 stability
We evaluated potential mechanisms for the decreased function of Tra1-L3733A. Since Tra1-L3733A interacted with the Gal4 activation domain and components of SAGA and NuA4 comparably to wild-type, the most straightforward explanation was its approximately fourfold reduced cellular concentration. Consistent with this model, similar growth defects are observed when the expression of wild-type Tra1 is decreased. Furthermore, intragenic suppressors of the L3733A mutation that increase activity restore Tra1 levels to a similar extent.
The reduced cellular concentration and altered proteolytic profile of Tra1-L3733A suggest a role for the FATC domain in maintaining the molecule’s three-dimensional structure. Spagnolo et al. (2006) found that the FATC domain of DNA-PKcs is involved in a conformational change that place it in close proximity to HEAT repeat sequences found toward the N-terminus. We speculate that the FATC domain of Tra1 may directly or indirectly, have a comparable role in determining conformation and that destabilizing this structure would result in enhanced sensitivity to proteolytic cleavage. In such a model suppression of L3733A by T3716A and N3677D may occur through reducing proteoytic degradation, perhaps through stabilizing necessary molecular interactions.
To directly compare the half-life of Tra1-L3733A with wild-type Tra1 we analyzed protein levels after cycloheximide arrest of translation. Conclusions from these experiments were limited because of minimal turnover of Tra1 after cycloheximide arrest. Because of this we have also considered possible effects of the L3733A mutation on aspects of the expression of Tra1. It is unlikely that translational control is affected by L3733A because this would not easily account for the conservation of L3733 across species and in the PIKK family, the observed second-site suppression, or the altered proteolytic pattern. Nor does the inserted GCA codon show a negative bias. The level of Tra1-L3733A is also not likely the result of altered transcription since gene profiling data indicates only marginally reduced expression of DED1 (the promoter for the plasmid copies of TRA1) and the phenotypes are observed when the L3733A mutation is expressed from its native promoter or the Met3 promoter (not shown). This being said, we cannot exclude contributions from more complex mechanisms: for example, the stabilization, processing or nuclear export of its mRNA transcript.
Dames et al. (2005) determined the structure of the Tor1 FATC domain in solution. They observed an extended α-helix that was interrupted by a hairpin followed by a candy cane-like loop for the terminal five residues, held in place by a disulfide bridge at positions that correspond to 3734 and 3741 of Tra1. In the Tor1 structure, the leucine equivalent to L3733 of Tra1 is positioned proximal to its terminal tryptophan where a hydrophobic interaction could potentially stabilize the conformation of the loop. We do not believe a similar structure exists for the FATC domain of Tra1 since the cysteines are not found in Tra1, nor is a glycine found within the loop that would facilitate the bend. In addition, while mutation of the terminal phenylalanine reduced growth, the phenotypic profile did not resemble tra1-L3733A. We also evaluated the possible importance of a hydrophobic patch created by extending the α-helix to the extreme C-terminus of Tra1 and the formation of a surface including L3733, F3740, and F3744. We conclude that if the extended helix is formed, the integrity of the hydrophobic patch is not likely important because mutation of F3740 to alanine did not result in obvious growth defects.
Integrity of the extreme C-terminus of Tra1 is essential for function
Being a 3744-residue protein, it seems unlikely that the C-terminus of Tra1 has a role in the innate folding of the protein. Rather, the reduced interaction of Tra1-G3745 with Esa1 and Spt7 support a model whereby the C-terminus is involved in protein–protein interactions necessary for function and stability of the protein. The extreme C-terminal sequences of Tra1 resemble the hydrophobic termini found for the interacting partners of PDZ domains (Tonikian et al. 2008) and the C-terminal phenylalanine of TraD, a protein required for bacterial F plasmid conjugation. Crystal structures of the TraD-TraM interaction show precise alignment of the TraD C-terminus with TraM (Lu et al. 2008). This interaction is disrupted by a glycine addition to TraD. The combination of the charged C-terminus on a hydrophobic residue creates a highly specific interaction site not otherwise found on the protein surface (Lu et al. 2008).
Sun et al. (2005) have shown that mutations within the FATC domain of ATM affect its interaction with Tip60. We analyzed whether a C-terminal fragment of Tra1 including PI3K and FATC domains is sufficient for interaction with Esa1, the yeast counterpart of Tip60, using both bacterially expressed proteins and two-hybrid analysis. In neither case was an interaction detected (not shown). This implies that other regions of Tra1 are also required for the interaction or that Esa1 is not a direct target of the FATC domain. Nevertheless, the finding that Tra1-G3745 associates poorly with Esa1 and Spt7 suggests that the C-terminus functions, at least in part, through protein–protein interactions.
tra1-G3745 and to a lesser extent tra1-L3733A act in a dominant negative fashion. This may seem inconsistent with their loss of function. We favor the idea that the dominant negative nature of these alleles is due to high levels of partial complexes and/or Tra1 proteolytic products since for tra1-G3745 and tra1-L3733A there was an inverse relationship between the extent to which the allele was dominant negative and its cellular concentration.
Recently, Tra1 has been found associated with a group of proteins including Rvb1, Rvb2, Asa1, Tel2, Tti1, and Tti2, all of which are essential (Shevchenko et al. 2008). While the exact composition and function of this ASTRA (for ASsembly of Tel, Rvb and Atm-like kinase) complex (Shevchenko et al. 2008) is unknown, we cannot exclude the possibility that some of the deficiencies associated with the FATC mutants result from changes in their association with these proteins.
References
Abraham RT (2004) PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair 3:883–887
Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman J, Côté J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18:5108–5119
Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Côté J (2008) Eaf1 is the platform for NuA4 molecular assembly that evoluntionarily links chromatin acetylation to ATP-dependent excahange of histone H2A variants. Mol Cell Biol 28:2257–2270
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1988) Current protocols in molecular biology. Greene/Wiley-Interscience, New York, NY
Babiarz JE, Halley JE, Rine J (2006) Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2AZ in Saccharomyces cerevisiae. Genes Dev 20:700–710
Barbaric S, Reinke H, Hörz W (2003) Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol 23:3468–3476
Beamish HJ, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, Jeggo P (2000) The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res 28:1506–1513
Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L (1992) Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251–265
Bhaumik SR, Green MR (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15:1935–1945
Bhaumik SR, Green MR (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22:7365–7371
Bhaumik SR, Raha T, Aiello DP, Green MR (2004) In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev 18:333–343
Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411–415
Bosotti R, Isacchi A, Sonnhammer EL (2000) FAT: a novel domain in PIK-related kinases. Trends Biochem Sci 25:225–227
Brandl CJ, Furlanetto AM, Martens JA, Hamilton KS (1993) Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J 12:5255–5265
Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333–2337
Burgess JR, Zhou H, Han J, Zhang Z (2010) A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell 37:469–480
Chen W, Struhl K (1988) Saturation mutagenesis of a yeast HIS3 “TATA element”: genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci USA 85:843–851
Choy JS, Kron SJ (2002) NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol 22:8215–8225
Cross FR (1997) ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647–653
Dames SA, Mulet JM, Rathgeb-Szabo K, Hall MN, Grzesiek S (2005) The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J Biol Chem 280:20558–20564
Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J (2004) Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16:979–990
Dudley AM, Rougeulle C, Winston F (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13:2940–2945
Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR 3rd, Berger SL, Workman JL (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol 19:6621–6631
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Evan GI, Lewis GK, Ramsay G, Bishop JM (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5:3610–3616
Fishburn J, Mohibullah N, Hahn S (2005) Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18:369–378
Gansheroff LF, Dollard C, Tan P, Winston F (1995) The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523–536
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425:737–741
Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25:31–42
Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11:1640–1650
Han M, Grunstein M (1988) Nucleosome loss activates yeast downstream promoters in vivo. Cell 55:1137–1145
Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104
Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17:2648–2663
Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang JQ (2001) Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet 29:206–211
Hoke SM, Genereaux J, Liang G, Brandl CJ (2008a) A conserved central region of yeast Ada2 regulates the histone acetyltransferase activity of Gcn5 and interacts with phospholipids. J Mol Biol 384:743–755
Hoke SM, Guzzo J, Andrews B, Brandl CJ (2008b) Systematic genetic array analysis links the Saccharomyces cerevisiae SAGA/SLIK and NuA4 component Tra1 to multiple cellular processes. BMC Genet 9:46
Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728
Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH (2000) Functional discovery via a compendium of expression profiles. Cell 102:109–126
Huisinga KL, Pugh BF (2004) A genome-wide housekeeping role for TFIID and a highly stress regulated role for SAGA in Saccharomyces cerevisiae. Mol Cell 13:573–585
Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL (2005) H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25:1162–1172
Jiang X, Sun Y, Chen S, Roy K, Price BD (2006) The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J Biol Chem 281:15741–15746
Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S (2006) The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell 17:4228–4236
Kohler A, Schneider M, Cabal CG, Nehrbass U, Hurt E (2008) Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10:707–715
Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien Z, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF (2003) A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12:1565–1576
Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, Emili A, Buratowski S, Hieter P, Greenblatt JF (2004) Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA 101:13513–13518
Larschan E, Winston F (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15:1946–1956
Larschan E, Winston F (2005) The Saccharomyces cerevisiae Srb8–Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol Cell Biol 24:114–123
Le Masson I, Yu DY, Jensen K, Chevalier A, Courbeyrette R, Boulard Y, Smith MM, Mann C (2003) Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol Cell Biol 23:6086–6102
Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28:327–334
Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD (2008) A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev 22:2062–2074
Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JNM (2008) Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol 70:89–99
Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633–643
Mao X, Hu Y, Liang C, Lu C (2002) MET3 promoter: a tightly regulated promoter and its application in construction of conditional lethal strain. Curr Microbiol 45:37–40
Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15:2227–2235
McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363–374
Millar CB, Xu F, Zhang K, Grunstein M (2006) Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev 20:711–722
Mitchell L, Lambert JP, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, Baetz K (2008) Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol 28:2244–2256
Mohibullah N, Hahn S (2008) Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22:2994–3006
Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S (2007) Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem 282:7799–7808
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z (2007) Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 26:5358–5372
Mutiu AI, Brandl CJ (2005) RNA isolation from yeast using silica matrices. J Biomol Tech 16:316–317
Mutiu AI, Hoke SM, Genereaux J, Hannam C, MacKenzie K, Jobin-Robitaille O, Guzzo J, Côté J, Andrews B, Haniford DB, Brandl CJ (2007a) Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast Tra1. Genetics 177:151–166
Mutiu AI, Hoke SM, Genereaux J, Liang G, Brandl CJ (2007b) The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol Genet Gen 277:491–506
Nourani AR, Utley RT, Allard S, Côté J (2004) Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J 23:2597–2607
Priestley A, Beamish HJ, Gell D, Amatucci AG, Muhlmann-Diaz MC, Singleton BK, Smith GC, Blunt T, Schalkwyk LC, Bedford JS, Jackson SP, Jeggo PA, Taccioli GE (1998) Molecular and biochemical characterization of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res 26:1965–1973
Reeves WM, Hahn S (2005) Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25:9092–9102
Reid JL, Iyer VR, Brown PO, Struhl K (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6:1297–1307
Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17:1030–1032
Rudra D, Zhao Y, Warner JR (2005) Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24:533–542
Ruiz-Garcia AB, Sendra R, Pamblanco M, Tordera V (1997) Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett 403:186–190
Saleh A, Lang V, Cook R, Brandl CJ (1997) Identification of native complexes containing the yeast coactivator/repressor proteins Ngg1/Ada3 and Ada2. J Biol Chem 272:5571–5578
Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR 3rd, Lees-Miller SP, Cole MD, Brandl CJ (1998) Tra1p is a component of the yeast AdaSpt transcriptional regulatory complexes. J Biol Chem 273:26559–26565
Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan NJ, Stewart AF (2008) Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol 9:R167
Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 22:511–519
Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotesrkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-Binding protein interaction. Mol Cell Biol 19:86–98
Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8:473–479
Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102:13182–13187
Sun Y, Xu Y, Roy K, Price BD (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 27:8502–8509
Takahashi T, Hara K, Inoue H, Kawa Y, Tokunaga C, Hidayat S, Yoshino K, Kuroda Y, Yonezawa K (2000) Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 5:765–775
Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, Held HA, Appleton BA, Evangelista M, Wu Y, Xin X, Chan AC, Seshagiri S, Sander C, Lasky LA, Boone C, Bader GD, Sidhu SS (2008) A specificity map for the PDZ domain family. PLoS Biol 6:e239
Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y (1998) The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell 2:869–875
Winzeler EA, Davis RW (1997) Functional analysis of the yeast genome. Curr Opin Genet Dev 7:771–776
Wu PY, Winston F (2002) Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol 22:5367–5379
Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA (2004) Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev 18:2024–2035
Acknowledgments
This work was supported by a Canadian Institutes of Health Research grant to CJB (MOP10845). SMTH was supported by a Frederick Banting and Charles Best Canada Graduate Scholarship sponsored by the CIHR, and AIM and SK by Western Graduate Scholarships. We would like to thank Mark Glover for communicating results prior to publication and suggesting the G3745 experiment, Fred Winston and Jacques Côté for yeast strains, and Megan Davey, David Edgell, Brian Shilton and David Haniford for comments on the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Braus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hoke, S.M.T., Irina Mutiu, A., Genereaux, J. et al. Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1. Curr Genet 56, 447–465 (2010). https://doi.org/10.1007/s00294-010-0313-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-010-0313-3