Abstract
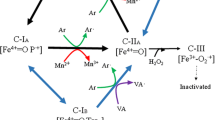
Lignin peroxidase (LiP) is the first enzyme connected to oxidative breakdown of the aromatic plant heteropolymer lignin and related xenobiotics. However, this extracellular enzyme has been described in only a few species of wood-decaying basidiomycetous fungi. The white rot basidiomycete Phlebia radiata 79 readily produces a versatile set of lignin-oxidizing enzymes including lignin and manganese peroxidases (LiPs and MnPs) and laccases. Here we describe genomic and primary structure of two new LiP-encoding genes, Pr-lip1 and Pr-lip4, and genomic characterization for isozyme LiP3/LIII of P. radiata, encoded by the gene depicted Pr-lip3. Pr-lip1 and Pr-lip4 code for 370- and 361-amino-acid long proteins beginning with 26- and 24-amino-acid secretion pre-propeptides, respectively. Translated LiP1 and LiP4 share the highest protein sequence identity (74 and 86%) with P. radiata LiP3, and 70% identity with the one deduced LiP from Bjerkandera adusta. The three P. radiata LiP sequences form a coherent phylogenetic cluster, which is further supported by similarities within gene organization interrupted by 11-introns. To find out the significance of LiP upon fungal growth on natural lignocellulose, such as wood, we studied ligninolytic gene expression on hardwood (milled alder) and softwood (spruce chips). All the LiP-encoding genes were expressed on wood with predominance of Pr-lip3 transcript abundance, in particular on spruce wood chips, where also time-dependent expression of the multiple lip genes was observed.





Similar content being viewed by others
References
Abbas A, Koc H, Liu F, Tien M (2005) Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Curr Genet 47:49–56
Blodig W, Smith AT, Doyle WA, Piontek K (2001) Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism. J Mol Biol 305:851–861
Bogan BW, Schoenike B, Lamar RT, Cullen D (1996) Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl Environ Microbiol 62:3697–3703
Broda P, Birch PRJ, Brooks PR, Sims PFG (1995) PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene-families. Appl Environ Microbiol 61:2358–2364
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Reporter 11:113–116
Cullen D (1997) Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol 53:273–289
Datta A, Bettermann A, Kirk TK (1991) Identification of a specific manganese peroxidase among ligninolytic enzymes secreted by Phanerochaete chrysosporium during wood decay. Appl Environ Microbiol 57:1453–1460
Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT (1998) Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry 37:15097–15105
Dresler-Nurmi A, Kaijalainen S, Lindström K, Hatakka A (1999) Grouping of lignin degrading corticioid fungi based on RFLP analysis of 18S rDNA and ITS regions. Mycol Res 103:990–996
Fedorov A, Merican AF, Gilbert W (2002) Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc Natl Acad Sci USA 99:16128–16133
Gilliland G, Perrin S, Bunn HF (1990) Competitive PCR for quantitation of mRNA. In: Innis MAea (ed) PCR protocols: a guide to methods and applications. Academic., San Diego, pp 60–69
Gold MH, Alic M (1993) Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev 57:602–622
Gold MH, Wariishi H, Valli K (1989) Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. In: Whitaker JR, Sonnet PE (eds) Biocatalysis in agricultural biotechnology vol 389: ACS symposium series. American Chemical Society, Washington, DC, pp 127–140
Gold MH, Youngs HL, Sollewijn Gelpke MD (2000) Manganese peroxidase. In: Sigel A, Sigel H (ed) Metalions in biological systems. Marcel Dekker Inc, New York, pp 559–586
Hakala TK, Maijala P, Konn J, Hatakka A (2004) Evaluation of novel wood-rotting polypores and corticioid fungi for the decay and biopulping of Norway spruce (Picea abies) wood. Enz Microb Technol 34:255–263
Hatakka A (1994) Lignin-modifying enzymes from selected white-rot fungi - production and role in lignin degradation. FEMS Microbiol Rev 13:125–135
Hatakka A, Buswell JA, Pirhonen TI, Uusi-Rauva AK (1983) Degradation of 14C-labelled lignins by white-rot fungi. In: Higuchi T, Chang H-M, Kirk TK (eds) Recent advances in lignin biodegradation research. UNI Publishers Co., Ltd., Tokyo, pp 176–187
Hatakka A, Lundell T, Tervilä-Wilo ALM, Brunow G (1991) Metabolism of nonphenolic beta-O-4 lignin model compounds by the white-rot fungus Phlebia radiata. Appl Microbiol Biotechnol 36:270–277
Hildén K, Martinez AT, Hatakka A, Lundell T (2005) The two manganese peroxidases Pr-MnP2 and Pr-MnP3 of Phlebia radiata, a lignin-degrading basidiomycete, are phylogenetically and structurally divergent. Fungal Genet Biol 42:403–419
Hofrichter M (2002) Lignin conversion by manganese peroxidase (MnP). Enz Microb Technol 30:454–466
Janse BJH, Gaskell J, Akhtar M, Cullen D (1998) Expression of Phanerochaete chrysosporium genes encoding lignin peroxidases, manganese peroxidases, and glyoxal oxidase in wood. Appl Environ Microbiol 64:3536–3538
Johansson T, Nyman PO (1996) A cluster of genes encoding major isozymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene 170:31–38
Johjima T, Itoh N, Kabuto M, Tokimura F, Nakagawa T, Wariishi H, Tanaka H (1999) Direct interaction of lignin and lignin peroxidase from Phanerochaete chrysosporium. Proc Natl Acad Sci USA 96:1989–1994
Kimura Y, Asada Y, Oka T, Kuwahara M (1991) Molecular analysis of a Bjerkandera adusta lignin peroxidase gene. Appl Microbiol Biotechnol 35:510–514
Kirk TK, Farrell RL (1987) Enzymatic “combustion”: the microbial degradation of lignin. Ann Rev Microbiol 41:465–505
Lamar RT, Schoenike B, Vanden Wymelenberg A, Stewart P, Dietrich DM, Cullen D (1995) Quantitation of fungal mRNAs in complex substrates by reverse transcription PCR and its application to Phanerochaete chrysosporium-colonized soil. Appl Environ Microbiol 61:2122–2126
Lugones LG, Scholtmeijer K, Klootwijk R, Wessels JG (1999) Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol 32:681–689
Lundell T, Leonowicz A, Rogalski J, Hatakka A (1990) Formation and action of lignin-modifying enzymes in cultures of Phlebia radiata supplemented with veratric acid. Appl Environ Microbiol 56:2623–2629
Lundell T, Schoemaker H, Hatakka A, Brunow G (1993) New mechanism of the Cα-Cβ cleavage in non-phenolic arylglycerol β-aryl ether lignin substructures catalyzed by lignin peroxidase. Holzforschung 47:219–224
Manubens A, Avila M, Canessa P, Vicuna R (2003) Differential regulation of genes encoding manganese peroxidase (MnP) in the basidiomycete Ceriporiopsis subvermispora. Curr Genet 43:433–438
Martínez AT (2002) Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb Technol 30:425–444
Martinez D, Larrondo LF, Putnam N, Sollewijn Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700
Mester T, Ambert-Balay K, Ciofi-Baffoni S, Banci L, Jones AD, Tien M (2001) Oxidation of tetrameric nonphenolic lignin model compound by lignin peroxidase. J Biol Chem 276:22985–22990
Moilanen A-M, Lundell T, Vares T, Hatakka A (1996) Manganese and malonate are individual regulators for the production of lignin and manganese peroxidase isozymes and in the degradation of lignin by Phlebia radiata. Appl Microbiol Biotechnol 45:792–799
Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, Mayama S (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42:275–283
Niku-Paavola M-L, Karhunen E, Kantelinen A, Viikari L, Lundell T, Hatakka A (1990) The effect of culture conditions on the production of lignin modifying enzymes by the white-rot fungus Phlebia radiata. J Biotechnol 13:211–221
Paszczynski A, Huynh V-B, Crawford R (1986) Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys 244:750–765
Piontek K, Glumoff T, Winterhalter K (1993) Low pH crystal structure of glyoxylated lignin peroxidase from Phanerochaete chrysosporium at 2.5 Å resolution. FEBS Lett 315:119–124
Poulos TL, Edwards SL, Wariishi H, Gold MH (1993) Crystallographic refinement of lignin peroxidase at 2 Å. J Biol Chem 268:4429–4440
Rajakumar S, Gaskell J, Cullen D, Lobos S, Karahanian E, Vicuna R (1996) lip-like genes in Phanerochaete sordida and Ceriporiopsis subvermispora, white-rot fungi with no detectable lignin peroxidase activity. Appl Environ Microbiol 62:2660–2663
Renganathan V, Miki K, Gold MH (1985) Multiple molecular forms of diarylpropane oxygenase, an H2O2-requiring, lignin-degrading enzyme from Phanerochaete chrysosporium. Arch Biochem Biophys 241:304–314
Ritch TJ, Nipper VJ, Akileswaran L, Smith AJ, Pribnow DG, Gold MH (1991) Lignin peroxidase from the basidiomycete Phanerochaete chrysosporium is synthesized as a preproenzyme. Gene 107:119–126
Saloheimo M, Barajas V, Niku-Paavola M-L, Knowles J (1989) A lignin peroxidase-encoding cDNA from the white-rot fungus Phlebia radiata: characterization and expression in Trichoderma reesei. Gene 85:343–351
Schoemaker H, Lundell T, Floris R, Glumoff T, Winterhalter K, Piontek K (1994) Do carbohydrates play a role in the lignin peroxidase cycle? Redox catalysis in the endergonic region of the driving force. Bioorg Med Chem 2:509–519
Stewart P, Cullen D (1999) Organization and differential regulation of a cluster of lignin peroxidase genes of Phanerochaete chrysosporium. J Bacteriol 181:3427–3432
Tien M, Kirk TK (1984) Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization and catalytic properties of a unique H2O2-requiring oxygenase. Proc Natl Acad Sci USA 81:2280–2284
Vares T, Kalsi M, Hatakka A (1995) Lignin peroxidases, manganese peroxidases, and other ligninolytic enzymes produced by Phlebia radiata during solid-state fermentation of wheat straw. Appl Environ Microbiol 61:3515–3520
Acknowledgments
The study was financially supported by the European Commission project QLK3-1999-00590 (PELAS), University of Helsinki (research grant #2108015 to T.L.), Academy of Finland (grant #53305 to the Center of Excellence on Microbial Resources; research grant #205027 to K.H.) and positions from the Viikki Graduate School of Biosciences (VGSB) to M.M. and T.H., which is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by U. Kück
Rights and permissions
About this article
Cite this article
Hildén, K.S., Mäkelä, M.R., Hakala, T.K. et al. Expression on wood, molecular cloning and characterization of three lignin peroxidase (LiP) encoding genes of the white rot fungus Phlebia radiata . Curr Genet 49, 97–105 (2006). https://doi.org/10.1007/s00294-005-0045-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-005-0045-y