Abstract
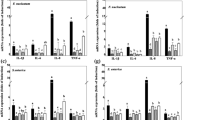
The present study aimed to isolate and identify the potential probiotic, pathobiont, and pathogenic microorganisms in the stool samples of 12 healthy individuals and evaluate their in vitro effects on cancer formation. A total of 83 strains were isolated from the stool samples and identified using MALDI-Biotyper. Fourteen of the isolates were identified as Candida spp., three isolates were identified as Cryptococcus neoformans, 55 isolates were identified as lactic acid bacteria, and the remaining isolates belonged to different 11 bacterial genera. Important microbial properties for cancer prevention and some probiotic properties were examined. All strains maintained their viability under acidic conditions and in media containing bile salts. Of the bacterial strains, 62.5% were resistant to ampicillin, chloramphenicol, gentamicin, erythromycin, kanamycin, penicillin, streptomycin, tetracycline, and vancomycin. All yeast strains were resistant to ketoconazole and susceptible to nystatin. The susceptibility of the strains to fluconazole, voriconazole, amphotericin B, and itraconazole varied. Fifty-nine percent of the strains produced EPS and 21.7% showed proteolytic activity (PA). Of the strains, 15.7% both produced exopolysaccharides (EPS) and had PA. The antioxidant activity (AOA) varied depending on the strains. The pathobiont and pathogenic microorganisms promoted tumor formation, while potential probiotic microorganisms had a suppressive effect on tumor formation (P > 0.01). One yeast (Candida kefyr MK17) and three lactic acid bacteria strains (Lacticaseibacillus paracasei MK73, Lactiplantibacillus plantarum MK55, Limosilactobacillus mucosae MK45) have superior potential thanks to their anticarcinogenic properties as well as tolerance to gastrointestinal tract conditions. Stool samples of each individual contain various potential probiotic, pathobiont, and pathogenic microorganisms.
Graphical Abstract




Similar content being viewed by others
Data Availability
Data will be made available on reasonable request.
Code Availability
Not applicable.
References
Vivarelli S, Salemi R, Candido S, Santagati FL, Stefani S, Torino F, Banna GL, Tonini G, Libra M (2019) Gut microbiota and cancer: From pathogenesis to therapy. Cancers 11:38. https://doi.org/10.3390/cancers11010038
Tsilimigras MCB, Fodor A, Jobin C (2017) Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol 2:17008. https://doi.org/10.1038/nmicrobiol.2017.8
Walter J, Maldonado-Gomez MX, Martinez I (2018) To engraft or not to engraft: An ecological framework for gut microbiome modulation with live microbes. Curr Opin Biotechnol 49:129–139. https://doi.org/10.1016/j.physbeh.2017.03.040
Sniffen JC, McFarland LV, Evans CT, Goldstein EJC (2018) Choosing an appropriate probiotic product for your patient: An evidence-based practical guide. PLoS ONE. https://doi.org/10.1371/JOURNAL.PONE.0209205
Lee YK, Salminen S (1995) The coming of age of probiotics. Trends Food Sci Technol 6:241–245. https://doi.org/10.1016/S0924-2244(00)89085-8
Kiliç GB, Karahan AG (2010) Identification of lactic acid bacteria isolated from the fecal samples of healthy humans and patients with dyspepsia, and determination of their pH, bile, and antibiotic tolerance properties. J Mol Microbiol Biotechnol 18:220–229. https://doi.org/10.1159/000319597
Sağlam H, Karahan AG (2021) Plasmid stability of potential probiotic Lactobacillus plantarum strains in artificial gastric juice, at elevated temperature, and in the presence of novobiocin and acriflavine. Arch Microbiol 203:183–191. https://doi.org/10.1007/s00203-020-02017-4
Wheeler ML, Limon JJ, Bar AS et al (2016) Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19:865–873. https://doi.org/10.1016/j.chom.2016.05.003
Boranbayeva T, Karahan AG, Tulemissova Z, Myktybayeva R, Özkaya S (2020) Properties of a new probiotic candidate and Lactobacterin-TK2 against diarrhea in calves. Probiotics Antimicrob Proteins 12:918–928. https://doi.org/10.1007/s12602-020-09649-4
Araya M, Morelli L, Reid G, Sanders ME, Stanton C, Pineiro M, Ben Embarek P (2002) Guidelines for the Evaluation of Probiotics in Food. FAO/WHO Work. Gr. Rep. Draft. Guidel. Eval. Probiotics Food, Jt. https://doi.org/10.1111/j.1469-0691.2012.03873
Ma F, Song Y, Sun M, Wang A, Jiang S, Mu G, Tuo Y (2021) Exopolysaccharide produced by Lactiplantibacillus plantarum-12 alleviates intestinal inflammation and colon cancer symptoms by modulating the gut microbiome and metabolites of C57BL/6 mice treated by azoxymethane/dextran sulfate sodium salt. Foods. https://doi.org/10.3390/foods10123060
Angelin J, Kavitha M (2020) Exopolysaccharides from probiotic bacteria and their health potential. Int J Biol Macromol 162:853–865. https://doi.org/10.1016/J.IJBIOMAC.2020.06.190
Hoffmann A, Kleniewska P, Pawliczak R (2021) Antioxidative activity of probiotics. Arch Med Sci 17:792–804
Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI (2011) Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci 108:6252–6257. https://doi.org/10.1073/pnas.1102938108
Syal P, Vohra A (2013) Probiotic potential of yeasts isolated from traditional Indian fermented foods. Int J Microbiol Res 5:390–398. https://doi.org/10.9735/0975-5276.5.2.390-398
CLSI (2016) Performance standards for antimicrobial susceptibility testing. 26th Edition, CLSI supplement M100S
CLSI (2009) Method for antifungal disk diffusion susceptibility testing of yeasts; Approved Guideline—Second Edition, CLSI Doc M44-A2, 29(17)
Amaretti A, Di Nunzio M, Pompei A, Raimondi S, Rossi M, Bordoni A (2013) Antioxidant properties of potentially probiotic bacteria: In vitro and in vivo activities. Appl Microbiol Biotechnol 97:809–817. https://doi.org/10.1007/s00253-012-4241-7
Son S, Lewis BA (2002) Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure−activity relationship. J Agric Food Chem 50:468–472. https://doi.org/10.1021/JF010830B
Liu WM, Scott KA, Dalgleish AG (2015) Supernatants of tumours treated with chemotherapy can alter tumour growth and development in vivo. Anticancer Res 35:1499–1508
Awaisheh SS, Obeidat MM, Al-Tamimi HJ et al (2016) In vitro cytotoxic activity of probiotic bacterial cell extracts against Caco-2 and HRT-18 colorectal cancer cells. Milchwissenshaft 69:27–31. https://doi.org/10.25968/MSI.2016.7
IBM Corp (2015) IBM SPSS Statistics for Windows
Wilson DA, Young S, Timm K et al (2017) Multicenter evaluation of the Bruker MALDI Biotyper CA system for the identification of clinically important bacteria and yeasts. Am J Clin Pathol 147:623–631. https://doi.org/10.1093/ajcp/aqw225
Kimura K, Nishio T, Mizoguchi C, Koizumi A (2010) Analysis of the composition of lactobacilli in humans. Biosci Microflora 29:47–50. https://doi.org/10.12938/bifidus.29.47
Forbes D, Ee L, Camer-Pesci P, Ward PB (2001) Faecal candida and diarrhoea. Arch Dis Child 84:328–331. https://doi.org/10.1136/adc.84.4.328
Biasoli MS, Tosello ME, Magaró HM (2002) Adherence of Candida strains isolated from the human gastrointestinal tract. Mycoses 45:465–469. https://doi.org/10.1046/j.1439-0507.2002.00793.x
Demicco EG, Kattapuram SV, Kradin RL, Rosenberg AE (2018) Infections of joints, synovium-lined structures, and soft tissue. In: Kradin RL (ed) Diagnostic Pathology of Infectious Disease. Elsevier, pp. 377–401 https://doi.org/10.1016/B978-0-323-44585-6.00015-1
Dufresne SF, Marr KA, Sydnor E et al (2014) Epidemiology of Candida kefyr in patients with hematologic malignancies. J Clin Microbiol 52:1830–1837. https://doi.org/10.1128/JCM.00131-14
Pettit RK, Repp KK, Hazen KC (2010) Temperature affects the susceptibility of Cryptococcus neoformans biofilms to antifungal agents. Med Mycol 48:421–426. https://doi.org/10.3109/13693780903136879
Yıldıran H, Başyiğit Kılıç G, Karahan Çakmakçı AG (2019) Characterization and comparison of yeasts from different sources for some probiotic properties and exopolysaccharide production. Food Sci Technol 39:646–653. https://doi.org/10.1590/fst.29818
Znaidi S, van Wijlick L, Hernández-Cervantes A et al (2018) Systematic gene overexpression in Candida albicans identifies a regulator of early adaptation to the mammalian gut. Cell Microbiol 20:1–21. https://doi.org/10.1111/cmi.12890
Urdaneta V, Casadesús J (2017) Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med 4:163. https://doi.org/10.3389/fmed.2017.00163
Mathur S, Singh R (2005) Antibiotic resistance in food lactic acid bacteria—a review. Int J Food Microbiol 105:281–295. https://doi.org/10.1016/j.ijfoodmicro.2005.03.008
Abriouel H, Casado Muñoz MDC, Lavilla Lerma L et al (2015) New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res Int 78:465–481. https://doi.org/10.1016/j.foodres.2015.09.016
Shridhar S, Dhanashree B (2019) Antibiotic susceptibility pattern and biofilm formation in clinical isolates of Enterococcus spp. Interdiscip Perspect Infect Dis 2019:7854968. https://doi.org/10.1155/2019/7854968
Zhang Z, Chen M, Yu Y, Pan S, Liu Y (2018) Antimicrobial susceptibility among gram-positive and gram-negative blood-borne pathogens collected between 2012–2016 as part of the Tigecycline Evaluation and Surveillance Trial. Antimicrob Resist Infect Control 7:152. https://doi.org/10.1186/s13756-018-0441-y
Gandhi TN, Patel MG, Jain MR (2015) Antifungal susceptibility of Candida against six antifungal drugs by disk diffusion method isolated from vulvovaginal candidiasis. Int J Cur Res Rev 7:20–25
Al-mamari A, Al-buryhi M, Al-heggami MA, Al-hag S (2014) Identify and sensitivity to antifungal drugs of Candida species causing vaginitis isolated from vulvovaginal infected patients in Sana’a city. Der Pharma Chem 6:336–342
Hazırolan G (2018) Investigation of in vitro susceptibility of non-albicans Candida species to fluconazole, itraconazole, voriconazole by reference broth microdilution method: Application of new species-specific clinical breakpoints and epidemiological cutoff values. Türk Mikrobiyol Cem Derg 48:38–44. https://doi.org/10.5222/tmcd.2018.038
Alp D, Kuleaşan H (2019) Adhesion mechanisms of lactic acid bacteria: conventional and novel approaches for testing. World J Microbiol Biotechnol 35:1–9. https://doi.org/10.1007/s11274-019-2730-x
Kšonžeková P, Bystrický P, Vlčková S et al (2016) Exopolysaccharides of Lactobacillus reuteri: Their influence on adherence of E. coli to epithelial cells and inflammatory response. Carbohydr Polym 141:10–19. https://doi.org/10.1016/j.carbpol.2015.12.037
Su LY, Shi YX, Yan MR, Xi Y, Su XL (2015) Anticancer bioactive peptides suppress human colorectal tumor cell growth and induce apoptosis via modulating the PARP-p53-Mcl-1 signaling pathway. Acta Pharmacol Sin 36:1514–1519. https://doi.org/10.1038/aps.2015.80
Lal P, Sharma D, Pruthi P, Pruthi V (2010) Exopolysaccharide analysis of biofilm-forming Candida albicans. J Appl Microbiol 109:128–136. https://doi.org/10.1111/j.1365-2672.2009.04634.x
Akçaǧlar S, Ener B, Töre O (2011) Acid proteinase enzyme activity in Candida albicans strains: A comparison of spectrophotometry and plate methods. Turkish J Biol 35:559–567. https://doi.org/10.3906/biy-1002-39
Lin M, Chang F (2000) Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig Dis Sci 45:1617–1622. https://doi.org/10.1023/a:1005577330695
Choi SS, Kim Y, Han KS, You S, Oh S, Kim SH (2006) Effects of Lactobacillus strains on cancer cell proliferation and oxidative stress in vitro. Lett Appl Microbiol 42:452–458. https://doi.org/10.1111/j.1472-765X.2006.01913.x
Abegg MA, Alabarse PVG, Schüller ÁK, Benfato MS (2012) Glutathione levels in and total antioxidant capacity of Candida sp. cells exposed to oxidative stress caused by hydrogen peroxide. Rev Soc Bras Med Trop 45:620–626. https://doi.org/10.1590/s0037-86822012000500015
Ceugniez A, Tourret M, Dussert E et al (2017) Interactions between Kluyveromyces marxianus from cheese origin and the intestinal symbiont Bacteroides thetaiotaomicron: Impressive antioxidative effects. LWT-Food Sci Technol 81:281–290. https://doi.org/10.1016/j.lwt.2017.03.056
Thirabunyanon M, Boonprasom P, Niamsup P (2009) Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol Lett. https://doi.org/10.1007/s10529-008-9902-3
Motevaseli E, Shirzad M, Akrami SM, Mousavi AS, Mirsalehian A, Modarressi MH (2013) Normal and tumour cervical cells respond differently to vaginal lactobacilli, independent of pH and lactate. J Med Microbiol 62:1065–1072. https://doi.org/10.1099/jmm.0.057521-0
Funding
This study was supported by the Süleyman Demirel University Scientific Research Projects Coordination Unit, Isparta, Turkey [Project Number: 4436-D2-15].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Münevver Kahraman, Aynur Gül Karahan, and Mustafa Ender Poyrazoğlu declare that they have no conflict of interest.
Ethical Approval
The trials were approved by the Akdeniz University Clinical Trials Ethics Committee (Protocol Number: 70904504/99).
Consent to Participate
Aynur Gül Karahan and Münevver Kahraman contributed to the study conception and design. Material preparation, data collection, and analyses were performed by Münevver Kahraman and Aynur Gül Karahan. The first draft of the manuscript was written by Münevver Kahraman and Aynur Gül Karahan, and all the authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Consent for Publication
Written informed consent was obtained from all the volunteers who participated in the study. Informed consent was documented in a written, signed and dated informed consent form.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kahraman, M., Karahan, A.G. & Terzioğlu, M.E. Characterization of Some Microorganisms from Human Stool Samples and Determination of Their Effects on CT26 Colorectal Carcinoma Cell Line. Curr Microbiol 79, 225 (2022). https://doi.org/10.1007/s00284-022-02915-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02915-4