Abstract
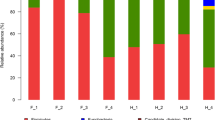
Fecal microbes play an important role in the survival and health of wild animals. Spotted hyena (Crocuta crocuta) is one of the representative carnivores in Africa. In this study, we examined the fecal microflora of spotted hyena by conducting high-throughput sequencing of the fecal microbial 16S rRNA gene V3–V4 high mutation region. The effects of age, sex, and feeding environment on the fecal microbiota of spotted hyenas were determined. The results showed that the core bacteria phyla of spotted hyenas fecal microbiota include Firmicutes (at an average relative abundance of 53.93%), Fusobacteria (19.56%), Bacteroidetes (11.40%), Actinobacteria (5.78%), and Proteobacteria (3.26%), etc. Age, gender, and feeding environment all had important effects on the fecal microbiota of spotted hyenas, among which feeding environment might be the most significant. The abundance of the Firmicutes in the adult group was significantly higher than that in the juvenile group, whereas the abundance of Fusobacteria, Bacteroidetes, and Proteobacteria were significantly lower than that in the juvenile group. The abundance of Lachnospiraceae and Ruminococcaceae in the female group was significantly higher than that in the male group. There were significant differences between the fecal microbial communities of Jinan group and Weihai group, and microbes from the phyla Firmicutes and Synergistetes were representative species associated with the difference.





Similar content being viewed by others
References
Heitlinger E, Ferreira SCM, Thierer D, Hofer H, East ML (2017) The intestinal eukaryotic and bacterial biome of spotted hyenas: the impact of social status and age on diversity and composition. Front Cell Infect Microbiol 7:262
Watts HE, Tanner JB, Lundrigan BL, Holekamp KE (2009) Post-weaning maternal effects and the evolution of female dominance in the spotted hyena. Proc Biol Sci 276(1665):2291–2298
Clements HS, Tambling CJ, Hayward MW, Kerley GI (2014) An objective approach to determining the weight ranges of prey preferred by and accessible to the five large african carnivores. PLoS ONE 9(7):e101054
Mills MGL, Hofer H (1998) Hyenas: status survey and conservation action plan. IUCN, Gland
Hayward MW (2006) Prey preferences of the spotted hyaena (Crocuta crocuta) and degree of dietary overlap with the lion (Pantera leo). J Zool 270:606–614
Yirga G, De Iongh HH, Leirs H, Gebrihiwot K, Deckers J, Bauer H (2012) Adaptability of large carnivores to changing anthropogenic food sources: diet change of spotted hyena (Crocuta crocuta) during Christian fasting period in Northern Ethiopia. J Anim Ecol 81(5):1052–1055
Wasimuddin MS, Melzheimer J, Thalwitzer S, Heinrich S, Wachter B, Sommer S (2017) Gut microbiomes of free-ranging and captive Namibian cheetahs: diversity, putative functions and occurrence of potential pathogens. Mol Ecol 26(20):5515–5527
Chen J, Zhang H, Wu X, Shang S, Yan J, Chen Y, Zhang H, Tang X (2017) Characterization of the gut microbiota in the golden takin (Budorcas taxicolor bedfordi). AMB Express 7(1):81
He J, Hai L, Orgoldol K, Yi L, Ming L, Guo F, Li G (2019) Ji R (2019) High-throughput sequencing reveals the gut microbiome of the bactrian camel in different ages. Curr Microbiol 76(7):810–817
Zhang HH, Chen L (2010) Phylogenetic analysis of 16S rRNA gene sequences reveals distal gut bacterial diversity in wild wolves (Canis lupus). Mol Biol Rep 37(8):4013–4022
Chen L, Zhang H, Liu G, Sha W (2016) First report on the bacterial diversity in the distal gut of dholes (Cuon alpinus) by using 16S rRNA gene sequences analysis. J Appl Genet 57(2):275–283
Han S, Guan Y, Dou H, Yang H, Yao M, Ge J, Feng L (2019) Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing. PeerJ 28(7):e6684
Garcia-Mazcorro JF, Lanerie DJ, Dowd SE, Paddock CG, Grützner N, Steiner JM, Ivanek R, Suchodolski JS (2011) Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol Ecol 78(3):542–554
Wu X, Zhang H, Chen J, Shang S, Wei Q, Yan J, Tu X (2016) Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3–V4 region of the 16S rRNA gene. Appl Microbiol Biotechnol 100(8):3577–3586
An C, Okamoto Y, Xu S, Eo KY, Kimura J, Yamamoto N (2017) Comparison of fecal microbiota of three captive carnivore species inhabiting Korea. J Vet Med Sci 79(3):542–546
Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (2006) The prokaryotes, vol 7. Springer, NewYork, pp 1016–1027
Marathe N, Shetty S, Lanjekar V, Ranade D, Shouche Y (2012) Changes in human gut flora with age: an Indian familial study. BMC Microbiol 12:222
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI (2012) Human gut microbiome viewed across age and geography. Nature 486(7402):222–227
Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, Ahn J (2015) Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS ONE 10(4):e0124599
Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H (2012) Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci USA 109(32):13034–13039
Wang J, Kalyan S, Steck N, Turner LM, Harr B, Künzel S, Vallier M, Häsler R, Franke A, Oberg HH, Ibrahim SM, Grassl GA, Kabelitz D, Baines JF (2015) Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat Commun 6:6440
McClelland EE, Smith JM (2011) Gender specific differences in the immune response to infection. Arch Immunol Ther Exp (Warsz) 59(3):203–213
Zhao L, Wang G, Siegel P, He C, Wang H, Zhao W, Zhai Z, Tian F, Zhao J, Zhang H (2013) Quantitative genetic background of the host influences gut microbiomes in chickens. Sci Rep 3(5):1970
Kruuk H (1972) The spotted hyena: a study of predation and social behavior. University of Chicago Press, Chicago
Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y (2014) Seasonal variation in human gut microbiome composition. PLoS ONE 9(3):e90731
Guan Y, Zhang H, Gao X, Shang S, Wu X, Chen J, Zhang W, Zhang W, Jiang M, Zhang B, Chen P (2016) Comparison of the bacterial communities in feces from wild versus housed sables (Martes zibellina) by high-throughput sequence analysis of the bacterial 16S rRNA gene. AMB Express 6(1):98
Zhao J, Yao Y, Li D, Xu H, Wu J, Wen A, Xie M, Ni Q, Zhang M, Peng G, Xu H (2018) Characterization of the gut microbiota in six geographical populations of chinese rhesus macaques (Macaca mulatta), implying an adaptation to high-altitude environment. Microb Ecol 76(2):565–577
Wu X, Zhang H, Chen J, Shang S, Yan J, Chen Y, Tang X, Zhang H (2017) Analysis and comparison of the wolf microbiome under different environmental factors using three different data of next generation sequencing. Sci Rep 7(1):11332
Grieneisen LE, Charpentier MJE, Alberts SC, Blekhman R, Bradburd G, Tung J, Archie EA (2019) Genes, geology and germs: gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc Biol Sci 286(1901):20190431
Acknowledgements
We would like to thank the staff of Jinan wildlife park, Weihai wildlife park and Linyi wildlife park for their assistance in the fecal samples collection. We would like to thank Editage (www.editage.cn) for English language editing.
Funding
This study was supported by grants from the National Natural Science Fund of China (No. 31400473), the Forestry Science and Technology Innovation Plan of Shandong province (LYCX07-2018-36), the Science and Technology Plan Project for Colleges and Universities in Shandong Province of China (J14LE16).
Author information
Authors and Affiliations
Contributions
LC conceived, designed, performed the experiments and analyzed the data; ML wrote the paper; YG, WS contributed materials; JZ, HD, WJ, SW modifies the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Rarefaction analysis of 12 adult spotted hyenas and 3 juvenile spotted hyena fecal microbiota. In the rarefaction curve, the abscissa represents the number of sequencing samples randomly extracted from a certain sample, and the ordinate represents the number of operational taxonomic units (OTUs) constructed on the basis of the number of sequences, which reflected the sequencing depth, and different samples are represented by differently colored curves (Png 111 kb )
284_2020_1914_MOESM2_ESM.png
Supplementary Figure 2 Anosim analyses of adult group and juvenile group (a), female group and male group (b), and Jinan group and Weihai group (c) based on Bray-Curtis distance.Anosim is used to investigate whether the intergroup differences are greater than the intragroup differences. The R value is between (-1, 1). An R value greater than 0 indicates that the intergroup difference is significant; an R value less than 0 indicates that the intragroup difference is greater than the intergroup difference. P values less than 0.05 indicated statistical significance. The ordinate represents the rank of the distance between the samples. The abscissa “Between” reflects intergroup findings and the other two reflect intragroup findings (Png 1,253 kb)
Rights and permissions
About this article
Cite this article
Chen, L., Liu, M., Zhu, J. et al. Age, Gender, and Feeding Environment Influence Fecal Microbial Diversity in Spotted Hyenas (Crocuta crocuta). Curr Microbiol 77, 1139–1149 (2020). https://doi.org/10.1007/s00284-020-01914-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01914-7