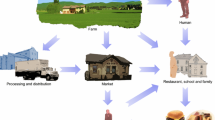
Abstract
The main Enterobacteriaceae habitat is the mammal gastrointestinal tract. In most cases, this group of species displays a symbiotic relationship with its hosts. However, some groups may be pathogenic to humans, such as Shiga toxin-producing Escherichia coli and enteroaggregative Escherichia coli. The presence of these groups represents a direct risk to consumers, and recent serotypes displaying the presence of pathogenic genes in both groups are a novel challenge for food production. Thus, microbiological control strategies presenting accurate detection methodologies are required. However, with the appearance of mutations among different species, knowledge, genetic monitoring, and bioinformatics techniques must be expanded. In addition, as a strategy to ensure safe products on an industrial scale, the monitoring by different techniques and fundamentals should be applied throughout the entire processing chain. Therefore, the aim of this review is to describe the pathogenesis mechanisms of different groups, mutant strain dispersion, and current and alternative epidemiological investigation methods.


Similar content being viewed by others
References
Kwong WK, Moran NA (2016) Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. https://doi.org/10.1038/nrmicro.2016.43
Santos ECCD, Castro VS, Cunha-Neto A et al (2018) Escherichia coli O26 and O113:H21 on carcasses and beef from a slaughterhouse located in Mato Grosso, Brazil. Foodborne Pathog Dis 15:653–659. https://doi.org/10.1089/fpd.2018.2431
Patzi-Vargas S, Zaidi MB, Perez-Martinez I et al (2015) Diarrheagenic Escherichia coli carrying supplementary virulence genes are an important cause of moderate to severe diarrhoeal disease in Mexico. PLoS Negl Trop Dis 9:e0003510. https://doi.org/10.1371/journal.pntd.0003510
Castro VS, Carvalho RCT, Conte-Junior CA, Figueiredo EES (2017) Shiga-toxin producing Escherichia coli: pathogenicity, supershedding, diagnostic methods, occurrence, and foodborne outbreaks. Compr Rev Food Sci Food Saf 16:1269–1280. https://doi.org/10.1111/1541-4337.12302
Oporto B, Esteban JI, Aduriz G et al (2008) Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in healthy cattle, sheep and swine herds in Northern Spain. Zoonoses Public Health 55:73–81. https://doi.org/10.1111/j.1863-2378.2007.01080.x
Bruyand M, Mariani-Kurkdjian P, Gouali M et al (2018) Hemolytic uremic syndrome due to Shiga toxin-producing Escherichia coli infection. Med Mal Infect 48:167–174. https://doi.org/10.1016/j.medmal.2017.09.012
Gyles CL (2007) Shiga toxin-producing Escherichia coli: an overview. J Anim Sci 85:E45–62. https://doi.org/10.2527/jas.2006-508
Jandhyala DM, Vanguri V, Boll EJ et al (2013) Shiga toxin-producing Escherichia coli O104:H4: an emerging pathogen with enhanced virulence. Infect Dis Clin North Am 27:631–649. https://doi.org/10.1016/j.idc.2013.05.002
Dudley EG, Abe C, Ghigo J-M et al (2006) An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect Immun 74:2102–2114. https://doi.org/10.1128/IAI.74.4.2102-2114.2006
Lavender HF, Jagnow JR, Clegg S (2004) Biofilm formation in vitro and virulence in vivo of mutants of Klebsiella pneumoniae. Infect Immun 72:4888–4890. https://doi.org/10.1128/IAI.72.8.4888-4890.2004
Rasko DA, Webster DR, Sahl JW et al (2011) Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N Engl J Med 365:709–717. https://doi.org/10.1056/NEJMoa1106920
Beutin L, Hammerl JA, Strauch E et al (2012) Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia. J Virol 86:10444–10455. https://doi.org/10.1128/JVI.00986-12
Nyholm O, Heinikainen S, Pelkonen S et al (2015) Hybrids of shigatoxigenic and enterotoxigenic Escherichia coli (STEC/ETEC) among human and animal isolates in Finland. Zoonoses Public Health 62:518–524. https://doi.org/10.1111/zph.12177
da Silva Santos AC, Gomes Romeiro F, Yukie Sassaki L, Rodrigues J (2015) Escherichia coli from Crohn’s disease patient displays virulence features of enteroinvasive (EIEC), enterohemorragic (EHEC), and enteroaggregative (EAEC) pathotypes. Gut Pathog 7(1):2. https://doi.org/10.1186/s13099-015-0050-8
Rajapaksha P, Elbourne A, Gangadoo S et al (2019) A review of methods for the detection of pathogenic microorganisms. Analyst 144:396–411. https://doi.org/10.1039/C8AN01488D
Nataro JP, Kaper JB, Robins-Browne R et al (1987) Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J 6:829–831
Hebbelstrup Jensen B, Olsen KEP, Struve C et al (2014) Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev 27:614–630. https://doi.org/10.1128/CMR.00112-13
Croxen MA, Law RJ, Scholz R et al (2013) Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. https://doi.org/10.1128/CMR.00022-13
Nataro JP, Kaper JB (1998) Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201
Jønsson R, Struve C, Boisen N et al (2015) Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect Immun 83:1396–1405. https://doi.org/10.1128/IAI.02820-14
Pereira ACM, Britto-Filho JD, José de Carvalho J et al (2008) Enteroaggregative Escherichia coli (EAEC) strains enter and survive within cultured intestinal epithelial cells. Microb Pathog 45:310–314. https://doi.org/10.1016/j.micpath.2008.07.001
Yang S-C, Lin C-H, Aljuffali IA, Fang J-Y (2017) Current pathogenic Escherichia coli foodborne outbreak cases and therapy development. Arch Microbiol 199:811–825. https://doi.org/10.1007/s00203-017-1393-y
Harrington SM, Dudley EG, Nataro JP (2006) Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett 254:12–18. https://doi.org/10.1111/j.1574-6968.2005.00005.x
Izquierdo M, Navarro-Garcia F, Nava-Acosta R et al (2014) Identification of cell surface-exposed proteins involved in the fimbria-mediated adherence of enteroaggregative Escherichia coli to intestinal cells. Infect Immun 82:1719–1724. https://doi.org/10.1128/IAI.01651-13
Jønsson R, Struve C, Boll EJ et al (2017) A Novel pAA Virulence plasmid encoding toxins and two distinct variants of the fimbriae of enteroaggregative Escherichia coli. Front Microbiol 8:263. https://doi.org/10.3389/fmicb.2017.00263
Fujiyama R, Nishi J, Imuta N et al (2008) The shf gene of a Shigella flexneri homologue on the virulent plasmid pAA2 of enteroaggregative Escherichia coli 042 is required for firm biofilm formation. Curr Microbiol 56:474–480. https://doi.org/10.1007/s00284-008-9115-y
Konowalchuk J, Speirs JI, Stavric S (1977) Vero response to a cytotoxin of Escherichia coli. Infect Immun 18:775–779
O’Brien AD, LaVeck GD, Thompson MR, Formal SB (1982) Production of Shigella dysenteriae type 1-like cytotoxin by Escherichia coli. J Infect Dis 146:763–769
Zweifel C, Fierz L, Cernela N et al (2017) Characteristics of Shiga toxin-producing Escherichia coli O157 in slaughtered reindeer from Northern Finland. J Food Prot 80:454–458. https://doi.org/10.4315/0362-028X.JFP-16-457
Bell BP, Goldoft M, Griffin PM et al (1994) A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 272:1349–1353
Karmali MA, Gannon V, Sargeant JM (2010) Verocytotoxin-producing Escherichia coli (VTEC). Vet Microbiol 140:360–370. https://doi.org/10.1016/j.vetmic.2009.04.011
De Schrijver K, Buvens G, Possé B, et al (2008) Outbreak of verocytotoxin-producing E. coli O145 and O26 infections associated with the consumption of ice cream produced at a farm, Belgium, 2007. Euro Surveill 13(7):9–10
Enache E, Mathusa EC, Elliott PH et al (2011) Thermal resistance parameters for Shiga toxin-producing Escherichia coli in apple juice. J Food Prot 74:1231–1237. https://doi.org/10.4315/0362-028X.JFP-10-488
Chaucheyras-Durand F, Durand H (2010) Probiotics in animal nutrition and health. Benef Microbes 1:3–9. https://doi.org/10.3920/BM2008.1002
Krause M, Barth H, Schmidt H (2018) Toxins of locus of enterocyte effacement-negative shiga toxin-producing Escherichia coli. Toxins. https://doi.org/10.3390/toxins10060241
Javadi M, Bouzari S, Oloomi M (2017) Horizontal gene transfer and the diversity of Escherichia coli. Recent Adv Physiol Pathog Biotechnol Appl. https://doi.org/10.5772/intechopen.68307
Worrall LJ, Bergeron JRC, Strynadka NCJ (2013) Chapter 14: type 3 secretion systems. In: Donnenberg MS (ed) Escherichia coli, 2nd edn. Academic Press, Boston, pp 417–450
Steyert SR, Sahl JW, Fraser CM et al (2012) Comparative genomics and stx phage characterization of LEE-negative shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol 2:133. https://doi.org/10.3389/fcimb.2012.00133
Buchholz U, Bernard H, Werber D et al (2011) German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. https://doi.org/10.1056/NEJMoa1106482
Bielaszewska M, Mellmann A, Zhang W, et al (2011) Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. https://doi.org/10.1016/S1473-3099
Hamm K, Barth SA, Stalb S et al (2016) Experimental Infection of Calves with Escherichia coli O104:H4 outbreak strain. Sci Rep 6:32812. https://doi.org/10.1038/srep32812
Navarro-Garcia F (2014) Escherichia coli O104:H4 pathogenesis: an enteroaggregative E. coli/Shiga Toxin-Producing E. coli explosive cocktail of high virulence. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.EHEC-0008-2013
Prager R, Lang C, Aurass P et al (2014) Two novel EHEC/EAEC hybrid strains isolated from human infections. PLoS ONE 9:e95379. https://doi.org/10.1371/journal.pone.0095379
Kim J, Oh K, Jeon S et al (2011) Escherichia coli O104:H4 from 2011 European outbreak and strain from South Korea. Emerg Infect Dis 17:1755–1756. https://doi.org/10.3201/eid1708.110879
Mora A, Herrrera A, López C et al (2011) Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int Microbiol 14:121–141. https://doi.org/10.2436/20.1501.01.142
Ferdous M, Zhou K, de Boer RF, et al (2015) Comprehensive characterization of Escherichia coli O104:H4 isolated from patients in the Netherlands. Front Microbiol 6:1348. https://doi.org/10.3389/fmicb.2015.01348
Carbonari CC, Deza N, Flores M et al (2014) First isolation of enteroaggregative Escherichia coli O104:H4 from a diarrhea case in Argentina. Rev Argent Microbiol 46:302–306. https://doi.org/10.1016/S0325-7541(14)70086-0
Dallman TJ, Chattaway MA, Cowley LA et al (2014) An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK. PLoS ONE 9:e98103. https://doi.org/10.1371/journal.pone.0098103
Iyoda S, Tamura K, Itoh K et al (2000) Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol Lett 191:7–10. https://doi.org/10.1111/j.1574-6968.2000.tb09311.x
Shridhar PB, Noll LW, Shi X et al (2016) Escherichia coli O104 in feedlot cattle feces: prevalence Isolation and Characterization. PLoS ONE 11:e0152101. https://doi.org/10.1371/journal.pone.0152101
Karmali MA (2018) Factors in the emergence of serious human infections associated with highly pathogenic strains of shiga toxin-producing Escherichia coli. Int J Med Microbiol 308:1067–1072. https://doi.org/10.1016/j.ijmm.2018.08.005
Doyle MP, Erickson MC (2006) Reducing the carriage of foodborne pathogens in livestock and poultry. Poult Sci 85:960–973. https://doi.org/10.1093/ps/85.6.960
Lenski RE, Wiser MJ, Ribeck N et al (2015) Sustained fitness gains and variability in fitness trajectories in the long-term evolution experiment with Escherichia coli. Proc Biol Sci 282:20152292. https://doi.org/10.1098/rspb.2015.2292
Aslani MM, Alikhani MY, Zavari A et al (2011) Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int J Infect Dis 15:e136–139. https://doi.org/10.1016/j.ijid.2010.10.002
Weintraub A (2007) Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J Med Microbiol 56:4–8. https://doi.org/10.1099/jmm.0.46930-0
Conrad CC, Stanford K, McAllister TA et al (2017) Competition during enrichment of pathogenic Escherichia coli may result in culture bias. FACETS 1:114–126. https://doi.org/10.1139/facets-2016-0007
Feng PCH, Jinneman K, Scheutz F, Monday SR (2011) Specificity of PCR and serological assays in the detection of Escherichia coli Shiga toxin subtypes. Appl Environ Microbiol 77:6699–6702. https://doi.org/10.1128/AEM.00370-11
EFSA (2008) Report from the Task Force on Zoonoses Data Collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. from food animals. EFSA J 6:141r. https://doi.org/10.2903/j.efsa.2008.141r
ISO (2012) ISO/TS‐13136:2012: microbiology of food and animal feed. real‐time polymerase chain reaction (PCR)—based method for the detection of food‐borne pathogens. Horizontal method for the detection of shiga toxin‐producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103 and O145 serogroups. Geneva, Switzerland: International Organization for Standardization.
EFSA Panel on Biological Hazards (2015) Scientific opinion on public health risks associated with enteroaggregative Escherichia coli (EAEC) as a food-borne pathogen. EFSA J 13(12):4330. https://doi.org/10.2903/j.efsa.2015.4330
Mellmann A, Harmsen D, Cummings CA et al (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6:e22751. https://doi.org/10.1371/journal.pone.0022751
Brzuszkiewicz E, Thürmer A, Schuldes J et al (2011) Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 193:883–891. https://doi.org/10.1007/s00203-011-0725-6
Parizad EG, Parizad EG, Valizadeh A (2016) The application of pulsed field gel electrophoresis in clinical studies. J Clin Diagn Res 10:DE01–DE04. https://doi.org/10.7860/JCDR/2016/15718.7043
Herschleb J, Ananiev G, Schwartz DC (2007) Pulsed-field gel electrophoresis. Nat Protoc 2:677–684. https://doi.org/10.1038/nprot.2007.94
Belén A, Pavón I, Maiden MCJ (2009) Multilocus sequence typing. Methods Mol Biol Clifton NJ 551:129–140. https://doi.org/10.1007/978-1-60327-999-4_11
Dallman TJ, Chattaway MA, Cowley LA et al (2014) An investigation of the diversity of trains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK. PLoS ONE 9(5):e98103. https://doi.org/10.1371/journal.pone.0098103
Tozzoli R, Grande L, Michelacci V et al (2014) Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli: a world in motion. Front Cell Infect Microbiol 4:80. https://doi.org/10.3389/fcimb.2014.00080
Pasquali F, Palma TF et al (2019) Whole genome sequencing based typing and characterisation of Shiga-toxin producing Escherichia coli strains belonging to O157 and O26 serotypes and isolated in dairy farms. Italian J Food Safety 7(4):7673. https://doi.org/10.4081/ijfs.2018.7673
Jenkins C (2018) Enteroaggregative Escherichia coli. Curr Top Microbiol Immunol 416:27–50. https://doi.org/10.1007/82_2018_105
Toro M, Cao G, Rump L, Nagaraja TG, Meng J, Gonzalez-Escalona N (2015) Genome sequences of 64 non-O157:H7 Shiga toxin-producing Escherichia coli strains. Genome Announc 3:e01067–e1115. https://doi.org/10.1128/genomeA.01067-15]
Ashton PM, Perry N, Ellis R, Petrovska L, Wain J, Grant KA, Jenkins C, Dallman TJ (2015) Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing. Peer J 3:e739. https://doi.org/10.7717/peerj.739
Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC et al (2012) Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci USA 109:3065–3070. https://doi.org/10.1073/pnas.1121491109
Roetzer A, Diel R, Kohl TA et al (2013) whole genome sequencing versus traditional genotyping for investigation of a mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. https://doi.org/10.1371/journal.pmed.1001387
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (Process: 310462/2018-5 and 311422/2016-0) and author Vinicius Castro thanks CNPq/Brazil for the PhD scholarship granted. Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (Grant Number E-26/203.049/2017 and E-26/201.859/2019) and author Ana Paletta thanks FAPERJ/Brazil for the scholarship granted.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Paletta, A.C.C., Castro, V.S. & Conte-Junior, C.A. Shiga Toxin-Producing and Enteroaggregative Escherichia coli in Animal, Foods, and Humans: Pathogenicity Mechanisms, Detection Methods, and Epidemiology. Curr Microbiol 77, 612–620 (2020). https://doi.org/10.1007/s00284-019-01842-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01842-1