Abstract
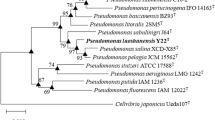
A Gram-stain-negative, rod-shaped bacterial strain JS15-10A1T was isolated from oil production water. Its optimum growth was observed at 37 °C, pH 7.0, and 3% (w/v) NaCl. The 16S rRNA gene sequence of strain JS15-10A1T showed the highest similarities with Pseudomonas parafulva CB-1T (97.6%) and P. fulva IAM 1529T (97.5%). In addition, phylogenetic analyses based on multilocus sequence analyses with concatenating 16S rRNA, gyrB, rpoD, and rpoB genes indicated that strain JS15-10A1T was a member of genus Pseudomonas but discriminated from other species. Furthermore, whole-genome analyses revealed that average nucleotide identities and in silico DNA-DNA hybridization values of strain JS15-10A1T against its closest relatives were all below 76.7% and 21.1%, respectively. The major cellular fatty acids of strain JS15-10A1T were summed feature 8 (C18:1ω7c/C18:1ω6c), C16:0, C12:0, C17:0 cyclo, summed feature 3 (C16:1ω7c/C16:1ω6c), C12:0 3-OH, C10:0 3-OH, and C19:0 cyclo ω8c. The predominant quinone was ubiquinone Q-9. The polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, an unknown amino-lipid, and two unidentified lipids. The genome DNA G + C content was 60.0 mol%. On the basis of phylogenetic, phenotypic, physiological, and chemotaxonomic analyses, it can be concluded that strain JS15-10A1T represents a novel species in genus Pseudomonas, for which the name Pseudomonas jilinensis sp. nov. is proposed. The type strain is JS15-10A1T (= CGMCC 1.16072T = LMG 30036T).

Similar content being viewed by others
Abbreviations
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- DPG:
-
Diphosphatidylglycerol
- AL:
-
Unknown amino-polar lipid
- L:
-
Unknown polar lipid
- ANI:
-
Average nucleotide identity
- MLSA:
-
Multilocus sequence analyses
- DDH:
-
DNA-DNA hybridization
- GGDC:
-
Genome-to-genome distance calculator
References
Migula W (1894) Über ein neues System der Bakterien. Arb Bakteriol Inst Karlsruhe 1:235–238
Lin SY, Hameed A, Liu YC, Hsu YH, Lai WA, Chen WM, Shen FT, Young CC (2013) Pseudomonas sagittaria sp. nov., a siderophore-producing bacterium isolated from oil-contaminated soil. Int J Syst Evol Microbiol 63:2410
Lang E, Burghartz M, Spring S, Swiderski J, Spröer C (2010) Pseudomonas benzenivorans sp. nov. and Pseudomonas saponiphila sp. nov., represented by xenobiotics degrading type strains. Curr Microbiol 60:85–91
Palleroni N (2005) Genus I Pseudomonas Migula 1894, 237AL. In: Brenner DJ, Krieg NR, Staley JT (eds) Bergey’s manual of systematic bacteriology. Springer, New York, pp 323–379
Moore ERB, Tindall BJ, Santos VAPMD, Pieper DH, Ramos JL, Palleroni NJ (2006) Nonmedical: Pseudomonas. Springer, New York, pp 646–703
Parte AC (2018) LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829
Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E (2015) Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214
Mulet M, Gomila M, Lemaitre B, Lalucat J, García-Valdés E (2012) Taxonomic characterization of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst Appl Microbiol 35:145–149
Peix A, Ramírez-Bahena MH, Velázquez E (2018) The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evol 57:106–116
Amoozegar MA, Shahinpei A, Sepahy AA, Makhdoumikakhki A, Seyedmahdi SS, Schumann P, Ventosa A (2014) Pseudomonas salegens sp. nov., a halophilic member of the genus Pseudomonas isolated from a wetland. Int J Syst Evol Microbiol 64:3565–3570
Anwar N, Abaydulla G, Zayadan B, Abdurahman M, Hamood B, Erkin R, Ismayil N, Rozahon M, Mamtimin H, Rahman E (2016) Pseudomonas populi sp. nov., an endophytic bacterium isolated from Populus euphratica. Int J Syst Evol Microbiol 66:1419–1425
Hwang CY, Zhang GI, Kang SH, Kim HJ, Cho BC (2009) Pseudomonas pelagia sp. nov., isolated from a culture of the Antarctic green alga Pyramimonas gelidicola. Int J Syst Evol Microbiol 59:3019–3024
Uchino M, Shida O, Uchimura T, Komagata K (2001) Recharacterization of Pseudomonas fulva Iizuka and Komagata 1963, and proposals of Pseudomonas parafulva sp. nov. and Pseudomonas cremoricolorata sp. nov. J Gen Appl Microbiol 47:247–261
Iizuka H, Komagata K (1963) New species of Pseudomonas belonged to fluorescent group (Studies on the microorganisms of cereal grains. Part V). J Agric Chem Soc Japan 37:137–141
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Biol 20:406–416
Dong XZ, Cai MY (2001) Determination of biochemical properties manual for the systematic identification of general bacteria. Science Press, Beijing, pp 370–398
Hildebrand DC, Palleroni NJ, Hendson M, Toth J, Johnson JL (1994) Pseudomonas flavescens sp. nov., isolated from walnut blight cankers. Int J Syst Bacteriol 44:410–415
Lin SY, Hameed A, Hung M, Liu YC, Hsu YH, Young LS, Young CC (2015) Pseudomonas matsuisoli sp. nov., isolated from a soil sample in Matsu Island (Taiwan). Int J Syst Evol Microbiol 65:902–909
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdésac E (2012) Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analyses approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol 35:455–464
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Saitou N, Imanishi T (1989) Relative efficiencies of the fitch-margoliash, maximum-parsimony, maximum-likelihood, minimum-evolution, and neighbor-joining methods of phylogenetic tree construction in obtaining the correct tree. Dental Press J Orthod 17(1):108–114
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki KI (1987) Lipid and cell-wall analyses in bacterial systematics. Methods Microbiol 19:161–207
Kew S, Banerjee T, Minihane AM, Finnegan YE, Williams CM, Calder PC (2003) Relation between the fatty acid composition of peripheral blood mononuclear cells and measures of immune cell function in healthy, free-living subjects aged 25–72 y. Am J Clin Nutr 77:1278–1286
Kates M (1986) Techniques of lipidology, 2nd edn. Elsevier, Amsterdam, pp 100–253
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Romanenko LA, Schumann P, Rohde M, Zhukova NV, Mikhailov VV, Stackebrandt E (2005) Marinobacter bryozoorum sp. nov., and Marinobacter sediminum sp. nov., novel bacteria from the marine environment. Int J Syst Evol Microbiol 55:143–148
Zhong ZP, Liu Y, Hou TT, Liu HC, Zhou YG, Wang F, Liu ZP (2015) Pseudomonas salina sp. nov., isolated from a salt lake. Int J Syst Evol Microbiol 65:2846–2851
Clark LL, Dajcs JJ, McLean CH, Bartell JG, Stroman DW (2006) Pseudomonas otitidis sp. nov., isolated from patients with otic infections. Int J Syst Evol Microbiol 56:709–714
Neubeck MV, Huptas C, Glück C, Krewinkel M, Stoeckel M, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M (2016) Pseudomonas helleri sp. nov. and Pseudomonas weihenstephanensis sp. nov., isolated from raw cow’s milk. Int J Syst Evol Microbiol 66:1163–1173
Acknowledgements
This work was supported by the Educational Commission of Anhui Province of China (Grant Nos. 2015zytz041, 2018jyxm1444), PetroChina Science and Technology Innovation Grant (Grant No. 2016D-5007–0701), the Independent Project Program of State Key Laboratory of Petroleum Pollution Control (Gene-guiding Isolation and Screening of n-alkane Degrading Bacteria; Grant No. PPCIP2017001), and CNPC Research Institute of Safety and Environmental Technology. The authors thank the Institute of Microbiology, Chinese Academy of Sciences, for providing the transmission electron microscopy facility.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank accession number for the 16S rRNA gene sequence of strain JS15-10A1T is KY486930. The complete genome sequence of strain JS15-10A1T is available from GenBank under accession number QJSA00000000. The digital protologue database (DPD) Taxon Number is TA00851.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, JW., Cai, M., Nie, Y. et al. Pseudomonas jilinensis sp. nov., Isolated from Oil Production Water of Jilin Oilfield in China. Curr Microbiol 77, 688–694 (2020). https://doi.org/10.1007/s00284-019-01798-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01798-2