Abstract
Background
The gut microbes of dairy cows are strongly associated with their health, but the relationship between milk production and the intestinal microbiota has seldom been studied. Thus, we explored the diversity of the intestinal microbiota during peak lactation of dairy cows.
Methods
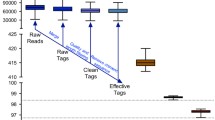
The intestinal microbiota of nine dairy cows at peak lactation was evaluated using the Pacific Biosciences single-molecule real-time (PacBio SMRT) sequencing approach.
Results
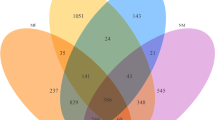
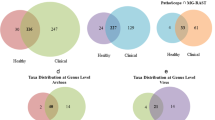
A total of 32,670 high-quality 16S rRNA gene sequences were obtained, belonging to 12 phyla, 59 families, 107 genera, and 162 species. Firmicutes (83%) were the dominant phylum, while Bacteroides (6.16%) was the dominant genus. All samples showed a high microbial diversity, with numerous genera of short chain fatty acid (SCFA)-producers. The proportion of SCFA producers was relatively high in relation to the identified core intestinal microbiota. Moreover, the predicted functional metagenome was heavily involved in energy metabolism.
Conclusions
This study provided novel insights into the link between the dairy cow gut microbiota and milk production.




Similar content being viewed by others
Availability of Data and Material
The sequence data reported have been deposited in the MG-RAST database under the project number mgp83332.
References
Godoyvitorino F, Goldfarb KC, Karaoz U, Leal S, Garciaamado MA et al. 2012. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J 6:531–541
Shabat SKB, Sasson G, Doronfaigenboim A, Durman T, Yaacoby S et al (2016) Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J 10(12):2958
Jr DJS (2010) Alternatives to conventional ileostomy—Mayo Clinic Proceedings. ISME J 4:1225–1235
Mayer M, Abenthum A, Matthes JM, Kleeberger D, Ege MJ et al (2012) Development and genetic influence of the rectal bacterial flora of newborn calves. Vet Microbiol 161:179
Brulc JM, Antonopoulos DA, Miller ME, Wilson MK, Yannarell AC et al. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 106:1948
Kim WI, Wonil K, Liu SY, Kinyon JM, Yoon KJ (2010) Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J Vet Diagn Investig 22:509–517
Jewell KA, Mccormick CA, Odt CL, Weimer PJ, Suen G (2015) Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710
Kohl KD, Weiss RB, Dale C, Dearing MD (2011) Diversity and novelty of the gut microbial community of an herbivorous rodent (Neotoma bryanti). Symbiosis 54:47
Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C (2011) Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol 8:36
Durso LM, Harhay GP, Smith TP, Bono JL, Desantis TZ et al (2010) Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microbiol 76:4858
Jami E, Israel A, Kotser A, Mizrahi I (2013) Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7(6):1069
Sasson G, Benshabat SK, Seroussi E, Doronfaigenboim A, Shterzer N et al (2017) Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. MBio 8:e00703-e007017
Plaizier JC, Li S, Tun HM, Khafipour E (2016) Nutritional models of experimentally-induced subacute ruminal acidosis (SARA) differ in their impact on rumen and hindgut bacterial communities in dairy cows. Front Microbiol 7:2128
Hou Q, Xu H, Zheng Y, Xi X, Kwok LY et al (2015) Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J Dairy Sci 98:8464–8472
Toma I, Siegel MO, Keiser J, Yakovleva A, Kim A et al (2014) Single-molecule long-read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol 52:3913–3921
Sutton JD (1985) Digestion and absorption of energy substrates in the lactating cow. J Dairy Sci 68:3376–3393
Gálfi P, Bokori J (1990) Feeding trial in pigs with a diet containing sodium n-butyrate. Acta Vet Hung 38:3–17
Xu H, Huang W, Hou Q, Kwok LY et al. (2017) The effects of probiotics administration on the milk production, milk components and fecal bacteria microbiota of dairy cows. Sci Bull 62(11)767–774
Pimentel PG, Reis RB, Neiva JNM, Coelho SG, Pinto AP (2017) Yield and composition of milk from dairy cows fed diets containing cashew nuts. Rev Cienc Agron 48(4):700–707
Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M et al (2009) Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol 56:80–87
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335
Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228
Cole JR, Chai B, Farris RJ, Wang Q, Kulamsyedmohideen AS et al (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35:169–172
Langille MGI, Zaneveld J, Caporaso JG, Mcdonald D, Dan K et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
O’Hara AM, Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7:688
Jeon SJ, Cunha F, Vieiraneto A, Bicalho RC, Lima S et al (2017) Blood as a route of transmission of uterine pathogens from the gut to the uterus in cows. Microbiome 5:109
Stein DR, Allen DT, Perry EB, Bruner JC, Gates KW et al (2006) Effects of feeding propionibacteria to dairy cows on milk yield, milk components, and reproduction 1. J Dairy Sci 89:111–125
Stella AV, Paratte R, Valnegri L, Cigalino G, Soncini G et al (2007) Effect of administration of live Saccharomyces cerevisiae on milk production, milk composition, blood metabolites, and faecal flora in early lactating dairy goats. Small Rumin Res 67:7–13
Fernando SC, Purvis HT, Najar FZ, Sukharnikov LO, Krehbiel CR et al (2010) Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol 76:7482
Kong Y, Teather R, Forster R (2010) Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol 74:612–622
Sadet-Bourgeteau S, Martin C, Morgavi DP (2010) Bacterial diversity dynamics in rumen epithelium of wethers fed forage and mixed concentrate forage diets. Vet Microbiol 146:98–104
Khafipour E, Li S, Plaizier JC, Krause DO (2009) Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol 75:7115–7124
Zhang J, Guo Z, Xue Z, Sun Z, Zhang M et al (2015) A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J 9:1979–1990
Zhao L, Zhang F, Ding X, Wu G, Lam YY et al (2018) Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156
Anderson KL (1995) Biochemical analysis of starch degradation by Ruminobacter amylophilus 70. Appl Environ Microbiol 61:1488
Qiao J, Kwok L, Zhang J, Gao P, Zheng Y et al (2015) Reduction of Lactobacillus in the milks of cows with subclinical mastitis. Benef Microbes 6:1–6
Chilliard Y, Ferlay A, Mansbridge RM, Doreau M, Agabriel J, Givens I (2000) Ruminant milk fat plasticity: nutritional control of saturated, polyunsaturated, trans and conjugated fatty acids. Annales De Zootech 49:181–205
de Oliveira MN, Jewell KA, Freitas FS, Benjamin LA, Tótola MR et al (2013) Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet Microbiol 164:307–314
Acknowledgements
We would like to express our gratitude to Professor Heping Zhang for his support, particularly for providing facilities to perform this work. Never forget why you started, and your mission can be accomplished. Funding was provided by National Natural Science Foundation of China.
Funding
This research was supported by the Science and Technology Major Project of Inner Mongolia Autonomous Region.
Author information
Authors and Affiliations
Contributions
TS and ZS designed the study. WL and JL collected samples. FZ and YL processed. HM and YW sequenced samples. WL and QH analyzed the data. WL, JY, and LK wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, W., Hou, Q., Wang, Y. et al. Analysis of the Gut Microbial Diversity of Dairy Cows During Peak Lactation by PacBio Single-Molecule Real-Time (SMRT) Sequencing. Curr Microbiol 75, 1316–1323 (2018). https://doi.org/10.1007/s00284-018-1526-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1526-9