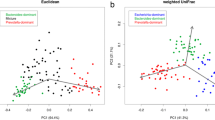
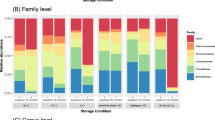
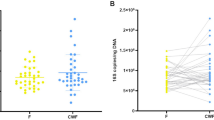
Abstract
Fecal sample collection is an important influential factor for DNA-based gut microbiota study. It is controversial whether the microbiome detected in fecal sample collected at one random day could fully represent the gut microbial community. The aim of the study is to figure out whether the use of fecal sample mixture collected at consecutive 5 days could more accurately represent gut microbial community. 1- and 5-day fecal samples were collected from 8 healthy adults and analyzed by 16S rRNA sequence. Our results indicated that both 1-day fecal samples and 5-day samples exhibited relatively high repeatability. The relative abundance of majority of bacterial taxa did not changed between 1-day fecal samples and 5-day fecal samples. However, the alpha diversity of 5-day fecal samples was higher than that of 1-day fecal samples. When the aims of studies are to analyze the relative abundance of specific OTUs among subjects, fecal samples collected at one day could be used. When microbial diversity is one of essential factors to be analyzed, the use of 5-day fecal samples may be more recommended.



Similar content being viewed by others
References
Ahn J, Sinha R, Pei Z et al (2013) Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105(24):1907–1911. https://doi.org/10.1093/jnci/djt300
Amato KR, Yeoman CJ, Kent A et al (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7(7):1344–1353. https://doi.org/10.1038/ismej.2013.16
Arumugam M, Raes J, Pelletier E et al (2011) Enterotypes of the human gut microbiome. Nature 473(7346):174–180. https://doi.org/10.1038/nature09944
Carroll IM, Ringel-Kulka T, Keku TO et al (2011) Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 301(5):G799-807. https://doi.org/10.1152/ajpgi.00154.2011
Chu H, Khosravi A, Kusumawardhani IP et al (2016) Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352(6289):1116–1120. https://doi.org/10.1126/science.aad9948
Clarke SF, Murphy EF, O’Sullivan O et al (2014) Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63(12):1913–1920. https://doi.org/10.1136/gutjnl-2013-306541
David LA, Materna AC, Friedman J et al (2014) Host lifestyle affects human microbiota on daily timescales. Genome Biol 15(7):R89. https://doi.org/10.1186/gb-2014-15-7-r89
David LA, Maurice CF, Carmody RN et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484):559–563. https://doi.org/10.1038/nature12820
Durban A, Abellan JJ, Jimenez-Hernandez N et al (2012) Daily follow-up of bacterial communities in the human gut reveals stable composition and host-specific patterns of interaction. FEMS Microbiol Ecol 81(2):427–437. https://doi.org/10.1111/j.1574-6941.2012.01368.x
Eckburg PB, Bik EM, Bernstein CN et al (2005) Diversity of the human intestinal microbial flora. Science 308(5728):1635–1638. https://doi.org/10.1126/science.1110591
Faith JJ, Guruge JL, Charbonneau M et al (2013) The long-term stability of the human gut microbiota. Science 341(6141):1237439. https://doi.org/10.1126/science.1237439
Falony G, Joossens M, Vieira-Silva S et al (2016) Population-level analysis of gut microbiome variation. Science 352(6285):560–564. https://doi.org/10.1126/science.aad3503
Flores R, Shi J, Fuhrman B et al (2012) Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med 10:253. https://doi.org/10.1186/1479-5876-10-253
Forslund K, Hildebrand F, Nielsen T et al (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528(7581):262–266. https://doi.org/10.1038/nature15766
Gorvitovskaia A, Holmes SP, Huse SM (2016) Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 4:15. https://doi.org/10.1186/s40168-016-0160-7
Halmos EP, Christophersen CT, Bird AR et al (2015) Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut 64(1):93–100. https://doi.org/10.1136/gutjnl-2014-307264
Hill CJ, Brown JR, Lynch DB et al (2016) Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome 4(1):19. https://doi.org/10.1186/s40168-016-0164-3
Jalanka-Tuovinen J, Salonen A, Nikkila J et al (2011) Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 6(7):e23035. https://doi.org/10.1371/journal.pone.0023035
Kennedy NA, Walker AW, Berry SH et al (2014) The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE 9(2):e88982. https://doi.org/10.1371/journal.pone.0088982
Koren O, Knights D, Gonzalez A et al (2013) A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9(1):e1002863. https://doi.org/10.1371/journal.pcbi.1002863
Le Chatelier E, Nielsen T, Qin J et al (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500(7464):541–546. https://doi.org/10.1038/nature12506
Man SM, Zhu Q, Zhu L et al (2015) Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 162(1):45–58. https://doi.org/10.1016/j.cell.2015.06.001
Noor SO, Ridgway K, Scovell L et al (2010) Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol 10:134. https://doi.org/10.1186/1471-230X-10-134
Rajilic-Stojanovic M, Heilig HG, Tims S et al (2012) Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. https://doi.org/10.1111/1462-2920.12023
Ridaura VK, Faith JJ, Rey FE et al (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341(6150):1241214. https://doi.org/10.1126/science.1241214
Scanlan PD, Shanahan F, O’Mahony C et al (2006) Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol 44(11):3980–3988. https://doi.org/10.1128/JCM.00312-06
Shaw AG, Sim K, Powell E et al. (2016) Latitude in sample handling and storage for infant faecal microbiota studies: the elephant in the room? Microbiome 4(1):40. https://doi.org/10.1186/s40168-016-0186-x
Sonnenburg ED, Sonnenburg JL (2014) Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab 20(5):779–786. https://doi.org/10.1016/j.cmet.2014.07.003
Vandeputte D, Falony G, Vieira-Silva S et al (2016) Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65(1):57–62. https://doi.org/10.1136/gutjnl-2015-309618
Vanhoutte T, Huys G, Brandt E et al (2004) Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol 48(3):437–446. https://doi.org/10.1016/j.femsec.2004.03.001
Wu GD, Chen J, Hoffmann C et al (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334(6052):105–108. https://doi.org/10.1126/science.1208344
Zoetendal EG, Rajilic-Stojanovic M, de Vos WM (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57(11):1605–1615. https://doi.org/10.1136/gut.2007.133603
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81472214), Zhejiang province key science and technology innovation team (2013TD13).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, T., Liu, R., Long, Y. et al. 1-Day or 5-Day Fecal Samples, Which One is More Beneficial to be Used for DNA-Based Gut Microbiota Study. Curr Microbiol 75, 288–295 (2018). https://doi.org/10.1007/s00284-017-1378-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1378-8