Abstract
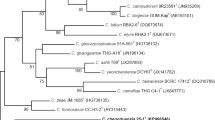
A novel bacterial strain, designated as CF21T, was isolated from the air of Ailuropoda melanoleuca enclosures in China. Cells were gram-negative, aerobic, non-motile, and rod shaped. Strain CF21T grew at 10–40 °C (optimum 28–30 °C) and pH 6.0–9.0 (optimum pH 7.0–8.0) and in the presence of NaCl concentrations ranging from 0.0 % (w/v) to 2.0 % (optimum 0.0–1.0 %). 16SrRNA gene sequence analysis indicated that strain CF21T belonged to genus Lysobacter within class Gammaproteobacteria and was most closely related to Luteimonas dalianensi OB44-3T (95.8 % similarity), Lysobacter ruishenii CTN-1T (95.1 %), Lysobacter spongiicola KMM329T (94.8 %), and Lysobacter daejeonensis GH1-9T (94.6 %). The genomic G+C DNA content was 68.72 mol%. Major cellular fatty acids of CF21T were iso-C16:0 (30.22 %), iso-C15:0 (25.70 %), and the sum of 10-methyl C16 : 0 and/or iso-C17 : 1ω9c (21.94 %). The prominent isoprenoid quinone was ubiquinone 8 (Q-8). Primary polar lipids included diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and an unknown phospholipid. DNA sequence relatedness between strain CF21T and L. ruishenii CTN-1T was 56 %, which was clearly below the 70 % threshold for prokaryotic species delineation. These analyses indicated that CF21T is a novel member of genus Lysobacter, for which the name Lysobacter chengduensis sp. nov. is proposed. The type strain is CF21T (=CGMCC1.15145T = DSM 100306T).

Similar content being viewed by others
References
Aslam Z, Yasir M, Jeon CO et al (2009) Lysobacter oryzae sp. nov., isolated from the rhizosphere of rice (Oryza sativa L.). Int J Syst Evol Microbiol 59:675–680
Bae HS, Im WT, Lee ST (2005) Lysobacter concretionis sp. nov., isolated from anaerobic granules in an upflow anaerobic sludge blanket reactor. Int J Syst Evol Microbiol 55:1155–1161
Buck JD (1982) Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
O’Hara CM (2006) Evaluation of the Phoenix 100 ID/AST system and NID Panel for identification of enterobacteriaceae, vibrionaceae, and commonly isolated nonenteric gram-negative bacilli. J Clin Microbiol 44:928–933
Choi JH, Seok JH, Cha JH et al (2014) Lysobacter panacisoli sp. nov., isolated from ginseng soil. Int J Syst Evol Microbiol 64:2193–2197
Christensen P, Cook FD (1978) Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol 28:367–393
Collins MD (1985) Isoprenoid quinone analysis in classification and identification. In: Minnikin MGDE (ed) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fukuda W, Kimura T, Araki S et al (2013) Lysobacter oligotrophicus sp. nov., isolated from an Antarctic freshwater lake in Antarctica. Int J Syst Evol Microbiol 63:3313–3318
Greisen K, Loeffelholz M, Purohit A et al (1994) PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol 32:335–351
Guindon S, Lethiec F, Duroux P et al (2005) PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33(Web Server issue):W557–W559
Islam MT, Hashidoko Y, Deora A et al (2005) Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne peronosporomycetes. Appl Environ Microbiol 71:3786–3796
Kim OS, Cho YJ, Lee K et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kluge AG, Farris JS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Liu M, Liu Y, Wang Y et al (2011) Lysobacter xinjiangensis sp. nov., a moderately thermotolerant and alkalitolerant bacterium isolated from a gamma-irradiated sand soil sample. Int J Syst Evol Microbiol 61:433–437
Luo G, Shi Z, Wang G (2012) Lysobacter arseniciresistens sp. nov., an arsenite-resistant bacterium isolated from iron-mined soil. Int J Syst Evol Microbiol 62:1659–1665
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Method Enzymol 12B:195–206
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Method 2:233–241
Park JH, Kim R, Aslam Z et al (2008) Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int J Syst Evol Microbiol 58:387–392
Romanenko LA, Uchino M, Tanaka N et al (2008) Lysobacter spongiicola sp. nov., isolated from a deep-sea sponge. Int J Syst Evol Microbiol 58:370–374
Saddler GS, Bradbury JF (2005) Family I. Xanthomonadaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematic bacteriology (the proteobacteria), part B (the gammaproteobacteria), vol 2, 2nd edn. Springer, New York, p 63
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Srinivasan S, Kim MK, Sathiyaraj G et al (2010) Lysobacter soli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 60:1543–1547
Stackebrandt E, Frederiksen W, Garrity GM et al (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Wang GL, Wang L, Chen HH et al (2011) Lysobacter ruishenii sp. nov., a chlorothalonil-degrading bacterium isolated from a long-term chlorothalonil-contaminated soil. Int J Syst Evol Microbiol 61:674–679
Weon HY, Kim BY, Baek YK et al (2006) Two novel species, Lysobacter daejeonensis sp. nov. and Lysobacter yangpyeongensis sp. nov., isolated from Korean greenhouse soils. Int J Syst Evol Microbiol 56:947–951
Ye XM, Chu CW, Shi C et al (2015) Lysobacter caeni sp. nov., isolated from the sludge of pesticide manufacturing factory. Int J Syst Evol Microbiol 65:845–850
Acknowledgments
We are grateful to Dr. Jian-Dong Jiang of Nanjing Agricultural University for providing L. ruishenii CTN-1T and L. daejeonensis GH1-9T. This work was supported by the National Science and Technology Support Project (2012BAC01B06) and Chengdu Giant Panda Breeding Research Foundation (CPF2010-06), China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2015_921_MOESM1_ESM.docx
Polar lipids analyses of strain Lysobacter chengduensis CF21T were carried out using a two-dimensional TLC method. PE, Phosphatidylethanolamine; PG, phosphatidylglycerol; DPG, diphosphatidylglycerol, and UPL, an unknown phospholipid. Supplementary material 1 (DOCX 612 kb)
Rights and permissions
About this article
Cite this article
Wen, C., Xi, L., She, R. et al. Lysobacter chengduensis sp. nov. Isolated from the Air of Captive Ailuropoda melanoleuca Enclosures in Chengdu, China. Curr Microbiol 72, 88–93 (2016). https://doi.org/10.1007/s00284-015-0921-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0921-8