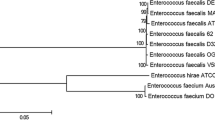
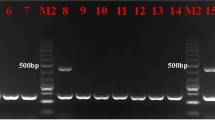
Abstract
Antibiotic-resistant opportunistic pathogens have become a serious concern in recent decades, as they are increasingly responsible for hospital-acquired infections. Here, we describe quinolone-resistant Delftia sp. strain 670, isolated from the sputum of a patient who died from severe pulmonary infection. The draft genome sequence of this strain was obtained by whole-genome shotgun sequencing, and was subjected to comparative genome analysis. Genome analysis revealed that one critical mutation (Ser83Ile in gyrA) might play a decisive role in quinolone resistance. The genome of Delftia sp. strain 670 contains both type II and type VI secretion systems, which were predicted to contribute to the virulence of the strain. Phylogenetic analysis, assimilation tests, and comparative genome analysis indicated that strain 670 differed from the four known Delftia species, suggesting this strain could represent a novel species. Although the study could not determine the strain 670 as the pathogen led to mortality, our findings also presented the pathogenic potential of Delftia species, and the increasing severity of antibiotic resistance among emerging opportunistic pathogens. The whole genome sequencing and comparative analysis improved our understanding of genome evolution in the genus Delftia, and provides the foundation for further study on drug resistance and virulence of Delftia strains.



Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Ambler R (1980) The Structure of beta-Lactamases. Philos Trans Royal Soc London B, Biol Sci 289(1036):321–331
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic acids research:gks1195
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10(1):421
Chen W-M, Lin Y-S, Sheu D-S, Sheu S-Y (2012) Delftia litopenaei sp. nov., a poly-β-hydroxybutyrate-accumulating bacterium isolated from a freshwater shrimp culture pond. Int J Syst Evol Microbiol 62(Pt 10):2315–2321
Chotikanatis K, Bäcker M, Rosas-Garcia G, Hammerschlag MR (2011) Recurrent intravascular-catheter-related bacteremia caused by Delftia acidovorans in a hemodialysis patient. J Clin Microbiol 49(9):3418–3421
Chun J, Lee J, Bae J, Kim M, Lee J-G, Shin S-Y, Kim YR, Lee K-H (2009) Delftia acidovorans isolated from the drainage in an immunocompetent patient with empyema. Tuberc Respir Dis 67(3):239–243
Del Mar Ojeda-Vargas M, Suárez-Alonso A, De Los Angeles Pérez-Cervantes M, Suárez-Gil E, Monzón-Moreno C (1999) Urinary tract infection associated with Comamonas acidovorans. Clin Microbiol Infect 5(7):443–444
Enoch D, Birkett C, Ludlam H (2007) Non-fermentative Gram-negative bacteria. Int J Antimicrob Agents 29:S33–S41
Hooper DC (2000) Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis 31(Supplement 2):S24–S28
Hooper DC (1999) Mechanisms of fluoroquinolone resistance. Drug Resist Updates 2(1):38–55
Horowitz H, Gilroy S, Feinstein S, Gilardi G (1990) Endocarditis associated with Comamonas acidovorans. J Clin Microbiol 28(1):143–145
Jørgensen NO, Brandt KK, Nybroe O, Hansen M (2009) Delftia lacustris sp. nov., a peptidoglycan-degrading bacterium from fresh water, and emended description of Delftia tsuruhatensis as a peptidoglycan-degrading bacterium. Int J Syst Evol Microbiol 59(9):2195–2199
Kashiwagi K, Tsuhako MH, Sakata K, Saisho T, Igarashi A, da Costa SOP, Igarashi K (1998) Relationship between spontaneous aminoglycoside resistance in Escherichia coli and a decrease in oligopeptide binding protein. J Bacteriol 180(20):5484–5488
Khan S, Sistla S, Dhodapkar R, Parija SC (2012) Fatal Delftia acidovorans infection in an immunocompetent patient with empyema. Asian Pacific J trop Biomed 2(11):923–924
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):0955–0964
Mustaev A, Malik M, Zhao X, Kurepina N, Luan G, Oppegard LM, Hiasa H, Marks KR, Kerns RJ, Berger JM (2014) Fluoroquinolone-Gyrase-DNA complexes two modes of drug binding. J Biol Chem 289(18):12300–12312
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34
Pennisi E (2010) Semiconductors inspire new sequencing technologies. Science 327(5970):1190–1191
Perla RJ, Knutson EL (2005) Delftia Acidovorans Bacteremia in an intravenous drug abuser. Am J Infect Dis 1(2):73
Preiswerk B, Ullrich S, Speich R, Bloemberg GV, Hombach M (2011) Human infection with Delftia tsuruhatensis isolated from a central venous catheter. J Med Microbiol 60(2):246–248
Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103(5):1528–1533
Pukatzki S, McAuley SB, Miyata ST (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol 12(1):11–17
Sandkvist M (2001) Type II secretion and pathogenesis. Infect Immun 69(6):3523–3535
Shigematsu T, Yumihara K, Ueda Y, Numaguchi M, Morimura S, Kida K (2003) Delftia tsuruhatensis sp. nov., a terephthalate-assimilating bacterium isolated from activated sludge. Int J Syst Evol Microbiol 53(5):1479–1483
Shin S, Choi J, Ko K (2012) Four cases of possible human infections with Delftia lacustris. Infection 40(6):709–712
Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK (2008) Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog 44(4):344–361
Tabak O, Mete B, Aydin S, Mandel NM, Otlu B, Ozaras R, Tabak F (2013) Port-related Delftia tsuruhatensis bacteremia in a patient with breast cancer. New Microbiol 36:199–201
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4(1):41
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Ciufo S, Li W (2013) Prokaryotic Genome Annotation Pipeline
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Von Graevenitz A (1995) Acinetobacter, Alcaligenes, Moraxella, and other nonfermentative gram-negative bacteria. Manual of clinical microbiology, 6th edn. American Society for Microbiology, Washington, DC, pp 520–532
Watts JL, Clinical, Institute LS (2008) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: approved standard. National Committee for Clinical Laboratory Standards
Wen A, Fegan M, Hayward C, Chakraborty S, Sly LI (1999) Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int J Syst Bacteriol 49(2):567–576
Wright GD (2011) Molecular mechanisms of antibiotic resistance. Chem Commun 47(14):4055–4061
Zheng J, Leung KY (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66(5):1192–1206
Acknowledgments
This research was supported by a grant from the National Hi-Tech Research and Development (863) Program of China (No. 2012AA022003 and No.2014AA021402), the China Mega-Project on Infectious Disease Prevention (Nos. 2013ZX10004605, 2011ZX10004001, 2013ZX10004607-004 and 2013ZX10004217-002-003) and the State Key Laboratory of Pathogen and BioSecurity Program (No. SKLPBS1113).
Conflict of interest
The authors declare that they have no conflict interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Huaixing Kang, Xiaomeng Xu and Kaifei Fu have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kang, H., Xu, X., Fu, K. et al. Characterization and Genomic Analysis of Quinolone-Resistant Delftia sp. 670 Isolated from a Patient Who Died from Severe Pneumonia. Curr Microbiol 71, 54–61 (2015). https://doi.org/10.1007/s00284-015-0818-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0818-6