Abstract
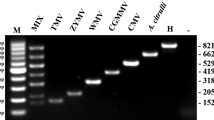
A specific and sensitive reverse transcriptase-nested polymerase chain reaction assay (RT-nPCR) was developed for the detection of Citrus tristeza virus (CTV) from naturally infected citrus samples. Two sets of primer pairs were designed by alignment of nucleotide sequences available in GenBank database for different genotypes of CTV. RT-nPCR reaction components and thermal cycling parameters were optimized and reaction conditions were standardized. Sequencing of the PCR products from direct and nested-PCR reactions confirmed the specificity of both primer pairs. Presence of CTV specific amplicons in asymptomatic samples which were collected from diseased orchards indicated the sensitivity of the test. As RT-nPCR technique, developed in the present study, is specific and efficient in detecting CTV, this could be envisioned for diagnostic applications and surveillance.


Similar content being viewed by others
References
Adkar-Purushothama CR, Gottravalli-Ramanayaka J, Sano T, Casati P, Bianco PA (2007) Are phytoplasmas the etiological agent of yellow leaf disease of Areca catechu in India?. Bull Insectol 60:161–162
Ananthakrishnan G, Venkataprasanna T, Roy A, Brlansky RH (2010) Characterization of the mixture of genotypes of a Citrus tristeza virus isolate by reverse transcription-quantitative real-time PCR. J Virol Methods 164:75–82
Bar-Joseph M, Marcus R, Lee RF (1989) The continuous challenge of Citrus tristeza virus control. Ann Rev Phytopathol 27:291–316
Brlansky RH, Lee RF, Garnsey SM (1988) In situ immunofluorescence for the detection of Citrus tristeza virus inclusion bodies. Plant Dis 72:1039–1041
Cambra M, Gorris MT, Marroquín C (2000) Incidence and epidemiology of Citrus tristeza virus in the Valencian Community of Spain. Virus Res 71:75–85
Cambra M, Olmos A, Gorris MT, Marroquíın C, Esteban O, Garnsey SM, Llauger R, Batista L, Peña I, Hermoso de Mendoza A (2000) Detection of Citrus tristeza virus by print capture and squash capture-PCR in plant tissue and single aphids. In: da Graća JV, Lee RF, Yokomi RK (eds) Proceedings of XIV IOCV conference, Riverside, CA, pp 42–49
Garnsey SM, Cambra M (1991) Enzyme-linked immunosorbent assay (ELISA) for citrus pathogens. In: Roistacher CN (ed) Graft transmissible diseases of citrus. Handbook for detection and diagnosis. FAO, Rome, pp 193–216
Gowda S, Satyanarayana T, Davis CL (2000) The p20 gene product of Citrus tristeza virus accumulates in the amorphous inclusion bodies. Virology 274:246–254
Hu JS, Li HP, Barry K (1995) Comparison of dot blot, ELISA, and RT-PCR assays for detection of two cucumber mosaic virus isolates infecting banana in Hawaii. Plant Dis 7:902–906
Huang Z, Rundell PA, Guan X (2004) Detection and isolate differentiation of Citrus tristeza virus in infected field trees based on reverse transcription polymerase chain reaction. Plant Dis 88:625–629
Hung TH, Wu ML, Su HJ (2000) A rapid method based on the onestep reverse transcriptase-polymerase chain reaction (RT-PCR) technique for detection of different strains of Citrus tristeza virus. J Phytopathol 148:469–475
Lee RF, Bar-Joseph M (2000) Tristeza. In: Timmer LW, Garnsey SM, Graham JH (eds) Compendium of citrus diseases, 2nd edn. American Phytopathological Society, St. Paul, MN, pp 61–63
Loconsole G, Saponari M, Savino V (2010) Development of real-time PCR based assays for simultaneous and improved detection of citrus viruses. Eur J Plant Pathol 128:251–259
Mackay I, Arden K, Nitsche A (2002) Real-time PCR in virology. Nucleic Acids Res 30:1292–1305
Marroquín C, Olmos A, Gorris MT (2004) Estimation of the number of aphids carrying Citrus tristeza virus that visit adult citrus trees. Virus Res 100:101–108
Mathews DM, Riley K, Dodds JA (1997) Comparison of detection methods for Citrus tristeza virus in field trees during months of nonoptimal titer. Plant Dis 81:525–529
Metha P, Brlansky RH, Gowda S (1997) Reverse-transcription polymerase chain reaction detection of Citrus tristeza virus in aphids. Plant Dis 81:1066–1069
Narvaez G, Skander BS, Ayllon MA (2000) A new procedure to differentiate Citrus tristeza virus isolates by hybridization with digoxigenin-labelled cDNA probes. J Virol Methods 85:83–92
Olmos A, Cambra M, Esteban O (1999) New device and method for capture, reverse transcription and nested PCR in a single closed-tube. Nucleic Acids Res 27:1564–1565
Powell CA, Pelosi RR, Rundell PA (2003) Prevalence of Citrus tristeza virus in Florida citrus nurseries and scion groves. HortScience 38:244–245
Roy A, Fayad A, Barthe G (2005) A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J Virol Methods 129:47–55
Saponari M, Manjunath K, Yokomi RK (2008) Quantitative detection of Citrus tristeza virus in citrus and aphids by real-time reverse transcription-PCR (TaqMan). J Virol Methods 147:43–53
Thompson JD, Higgins DG, Gibson TJ (1994) Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Yokomi RK, Saponari M, Sieburth PJ (2010) Rapid differentiation and identification of potential severe strains of Citrus tristeza virus by real-time reverse transcription-polymerase chain reaction assays. Phytopathology 100:319–327
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adkar-Purushothama, C.R., Maheshwar, P.K., Sano, T. et al. A Sensitive and Reliable RT-Nested PCR Assay for Detection of Citrus tristeza Virus from Naturally Infected Citrus Plants. Curr Microbiol 62, 1455–1459 (2011). https://doi.org/10.1007/s00284-011-9883-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-9883-7