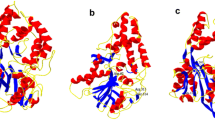
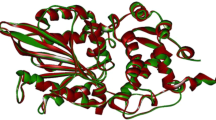
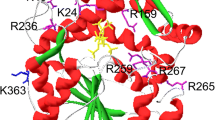
Abstract
In order to improve the thermostability of Escherichia coli AppA phytase, Error-prone PCR was used to randomize mutagenesis appA gene, and a gene mutation library was constructed. A mutant I408L was selected from the library by the method of high-throughput screening with 4-methyl-umbelliferylphosphate (4-MUP). The appA gene of the mutant was cloned and expressed in E. coli Origami (DE3). The recombinant protein was purified by Ni-affinity chromatography, and the enzymatic features were analyzed. The results indicated that AppA phytase activities of mutant I408L and wild-type (WT) strain remained at 51.3 and 28%, respectively, after treatment at 85°C for 5 min. It means that the thermostability enhancement of AppA phytase I408L was 23.3% more as compared with WT. The K m of both phytase were 0.18 and 0.25 mM, respectively, which indicated that the catalyzing efficiency of I408L was improved. AppA phytase of mutant I408L showed a significant enhancement against trypsin, which was nearly three times compared with WT. In addition, AppA phytase of mutant could be activated by Mg2+ and Mn2+; in contrast, it could be inhibited by Ca2+, Co2+, Cu2+, and K+ in varying degrees, and the enzymatic activity was almost lost the presence of Fe3+ and Zn2+. It appears that screening thermotolerant phytase of E. coli by high throughput screening with a fluorescence substrate is a fast, simple, and effective method. The mutant I408L obtained in this study could be used for the large-scale commercial production of phytase.




Similar content being viewed by others
References
Haefner S, Knietsch A, Scholten E, Braun J, Lohscheidt M, Zelder O (2005) Biotechnological production and applications of phytases. Appl Microbiol Biotechnol 68(5):588–597
Selle PH, Aaron J, Cowieson AJ, Ravindran V (2009) Consequences of calcium interactions with phytate and phytase for poultry and pigs. Livestock Sci 124(1–3):126–141
Lei XG, Stahl CH (2001) Biotechnological development of effective phytases for mineral nutrition and environmental protection. Appl Microbiol Biotechnol 57(4):474–481
Wyss M, Brugger R, Kronenberger A, Remy R, Fimbel R, Oesterhelt G, Lehmann M, van Loon AP (1999) Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl Environ Microbiol 65(2):367–373
Luo H, Huang H, Yang P, Wang Y, Yuan T, Wu N, Yao B, Fan Y (2007) A novel phytase appA from Citrobacter amalonaticus CGMCC 1696: gene cloning and overexpression in Pichia pastoris. Curr Microbiol 55(3):185–192
Chen KQ, Arnold FH (1991) Enzyme engineering for nonaqueous solvents: random mutagenesis to enhance activity of subtilisin E in polar organic media. Biotechnology 9(11):1073–1077
Stemmer WPC (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370(4):389–391
Fisch I, Kontermann RE, Finnern R, Hartley O, Soler-Gonzalez AS, Griffiths AD, Winter G (1996) A strategy of exon shuffling for making large peptide repertoires displayed on filamentous bacteriophage. Proc Natl Acad Sci USA 93(15):7761–7766
Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol 16(3):258–261
Rodriguez E, Wood ZA, Karplus PA, Lei XG (2000) Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch Biochem Biophys 382(1):105–112
Garrett JB, Kretz KA, Donoghue EO, Kerovuo J, Kim W, Barton NR, Hazlewood GP, Short JM, Robertson DE, Gray KA (2004) Enhancing the thermal tolerance and gastric performance of a microbial phytase for use as a phosphate-mobilizing monogastric-feed supplement. Appl Environ Microbiol 70(5):3041–3046
Kim MS, Lei XG (2008) Enhancing thermostability of Escherichia coli phytase AppA2 by error-prone PCR. Appl Microbiol Biotechnol 79(1):69–75
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Kikuchi M, Ohnishi K, Harayama S (1999) An effective family shuffling method using single-stranded DNA. Gene 243(1–2):133–137
Declerck N, Machius M, Wiegand G, Huber R, Gaillardin C (2000) Probing structural determinants specifying high thermostability in Bacillus licheniformis α-amylase. J Mol Biol 301(4):1041–1057
Giver L, Gershenson A, Freskgard PO, Arnold FH (1998) Directed evolution of a thermostable esterase. Proc Natl Acad Sci USA 95(22):12809–12813
Zhao H, Arnold FH (1999) Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng 12(1):47–53
Toyama H, Toyama N (1999) Construction of cellulase hyperproducing strains derived from polyploids of Trichoderma reesei. Microbios 100(395):7–18
Shibuya H, Kaneko S, Hayashi K (2000) Enhancement of the thermostability and hydrolytic activity of xylanase by random gene shuffling. Biochem J 349(2):651–656
Ragon M, Neugnot-Roux V, Chemardin P, Moulin G, Boze H (2008) Molecular gene cloning and overexpression of the phytase from Debaryomyces castellii CBS 2923. Protein Expr Purif 58(2):275–283
Li X, Liu Z, Chi Z, Li J, Wang X (2009) Molecular cloning, characterization, and expression of the phytase gene from marine yeast Kodamaea ohmeri BG3. Mycol Res 113(1):24–32
Ullah AH, Gibson DM (1988) Purification and characterization of acid phosphatase from cotyledons of germinating soybean seeds. Arch Biochem Biophys 260(2):514–520
Casey A, Walsh G (2003) Purification and characterization of extracellular phytase from Aspergillus niger ATCC 9142. Bioresour Technol 86(2):183–188
Singh B, Satyanarayana T (2008) Improved phytase production by a thermophilic mould Sporotrichum thermophile in submerged fermentation due to statistical optimization. Bioresour Technol 99(4):824–830
Wodzinski RJ, Ullah AH (1996) Phytase. Adv Appl Microbiol 42:263–302
Rodriguez E, Porres JM, Han Y, Lei XG (1999) Different sensitivity of recombinant Aspergillus niger phytase (r-PhyA) and Escherichia coli pH 2.5 acid phosphatase (r-AppA) to trypsin and pepsin in vitro. Arch Biochem Biophys 365(2):262–267
Dharmsthiti S, Chalermpornpaisarn S, Kiatiyajarn M, Chanpokapaiboon A, Klongsithidej Y, Techawiparut J (2005) Phytase production from Pseudomonas putida harbouring Escherichia coli appA. Process Biochem 40(2):789–793
Acknowledgments
This study was supported by The National Natural Science Foundation of China (30871321, 30771312, and 30971817), The National Special Basic Research Projects of China (SB2007FY400-4), and The National Basic Research Program of China (2009CB125910).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, W., Qiao, D., Huang, M. et al. Modifying Thermostability of appA from Escherichia coli . Curr Microbiol 61, 267–273 (2010). https://doi.org/10.1007/s00284-010-9606-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9606-5