Abstract
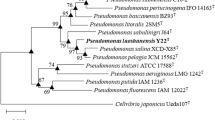
Isolate RS1T isolated from used metalworking fluid was found to be a Gram-negative, motile, and non-spore forming rod. Based on phylogenetic analyses with 16S rRNA, isolate RS1T was placed into the mendocina sublineage of Pseudomonas. The major whole cell fatty acids were C18:1ω7c (32.6%), C16:0 (25.5%), and C15:0 ISO 2OH/C16:1ω7c (14.4%). The sequence similarities of isolate RS1T based on gyrB and rpoD genes were 98.9 and 98.0% with Pseudomonas pseudoalcaligenes, and 98.5 and 98.1% with Pseudomonas oleovorans, respectively. The ribotyping pattern showed a 0.60 similarity with P. oleovorans ATCC 8062T and 0.63 with P. pseudoalcaligenes ATCC17440T. The DNA G + C content of isolate RS1T was 62.2 mol.%. The DNA–DNA relatedness was 73.0% with P. oleovorans ATCC 8062T and 79.1% with P. pseudoalcaligenes ATCC 17440T. On the basis of morphological, biochemical, and molecular studies, isolate RS1T is considered to represent a new subspecies of P. oleovorans. Furthermore, based on the DNA–DNA relatedness (>70%), chemotaxonomic, and molecular profile, P. pseudoalcaligenes ATCC 17440T and P. oleovorans ATCC 8062T should be united under the same name; according to the rules of priority, P. oleovorans, the first described species, is the earlier synonym and P. pseudoalcaligenes is the later synonym. As a consequence, the division of the species P. oleovorans into two novel subspecies is proposed: P. oleovorans subsp. oleovorans subsp. nov. (type strain ATCC 8062T = DSM 1045T = NCIB 6576T), P. oleovorans subsp. lubricantis subsp. nov. (type strain RS1T = ATCC BAA-1494T = DSM 21016T).




Similar content being viewed by others
References
Barrow GL, Feltham RKA (2004) Cowan and Steel’s manual for the identification of medical bacteria, 3rd edn. Cambridge University Press, Cambridge
Botes J, Williamson G, Sinicka V et al (2003) Genomic typing of Pseudomonas aeruginosa isolates by comparison of Riboprinting and PFGE: correlation of experimental results with those predicted from the complete genome sequence of isolate PAO1. J Microbiol Methods 55:231–240
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Gavini F, Holmes B, Izard D et al (1989) Numerical taxonomy of Pseudomonas alcaligenes, P. pseudoalcaligenes, P. mendocina, P. stutzeri, and related bacteria. Int J Syst Bacteriol 39:135–144
Gillis M, Vandamme P, De Vos P (2005) Polyphasic taxonomy. In: Brenner DJ, Kreig NR, Staley JT et al (eds) Bergey’s manual of systematic bacteriology. Springer, New York, pp 43–48
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kimura M (1980) A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lee M, Chandler AC (1941) A study of the nature, growth and control of bacteria in cutting compounds. J Bacteriol 41:373–386
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Palleroni NJ, Moore ERB (2004) Taxonomy of Pseudomonads: experimental approaches. In: Ramos JL (ed) Pseudomonas: genomics. Life style and Molecular Architecture, Birkhäuser, p 15
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomy study. J Appl Bacteriol 57:381–394
Sawada H, Suzuki F, Matsuda I et al (1999) Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J Mol Evol 49:627–644
Stackebrandt E, Frederiksen W, Garrity GM et al (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047
Sistanich D, Sherriff M (2005) The RiboPrinter® characterization system: an overview of automated ribotyping. In: Miller MJ (ed) Encyclopedia of rapid microbiological methods, vol 3. DHI Publishing, PDA Bethesda, MD
Thompson JD, Gibson TJ, Plewniak F et al (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Wayne LG, Brenner DJ, Colwell RR et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Yamamoto S, Harayama S (1995) PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol 61:1104–1109
Yamamoto S, Harayama S (1998) Phylogenetic relationships of Pseudomonas putida strains deduced from the nucleotide sequences of gyrB, rpoD and 16s rRNA genes. Int J Syst Evol Microbiol 48:813–819
Yamamoto S, Kasai H, Arnold DL et al (2000) Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394
Acknowledgments
We would like to extend our sincere thanks to Dr. Jeff Landgraf and Joseph Leykam of RTSF at Michigan State University, Michigan, USA for the determination of % G + C. We would also like to thank Dr. Ellen Dickstein at University of Florida, Gainesville, Florida, USA for FAME analysis, Dr. Stacie Frye at Bacterial Barcodes, Inc., Athens, Georgia, USA for Rep-PCR analysis and Bettina Sträubler at DSMZ for her excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saha, R., Spröer, C., Beck, B. et al. Pseudomonas oleovorans subsp. lubricantis subsp. nov., and Reclassification of Pseudomonas pseudoalcaligenes ATCC 17440T as Later Synonym of Pseudomonas oleovorans ATCC 8062T. Curr Microbiol 60, 294–300 (2010). https://doi.org/10.1007/s00284-009-9540-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9540-6