Abstract
Purpose
Embryonal rhabdomyosarcoma (eRMS) is the most common type of rhabdomyosarcoma in children. eRMS is characterized by malignant skeletal muscle cells driven by hyperactivation of several oncogenic pathways including the MYC pathway. Targeting MYC in cancer has been extremely challenging. Recently, we have demonstrated that the heart failure drug, proscillaridin A, produced anticancer effects with specificity toward MYC expressing leukemia cells. We also reported that decitabine, a hypomethylating drug, synergizes with proscillaridin A in colon cancer cells. Here, we investigated whether proscillaridin A exhibits epigenetic and anticancer activity against eRMS RD cells, overexpressing MYC oncogene, and its combination with decitabine.
Methods
We investigated the anticancer effects of proscillaridin A in eRMS RD cells in vitro. In response to drug treatment, we measured growth inhibition, cell cycle arrest, loss of clonogenicity and self-renewal capacity. We further evaluated the impact of proscillaridin A on MYC expression and its downstream transcriptomic effects by RNA sequencing. Then, we measured protein expression of epigenetic regulators and their associated chromatin post-translational modifications in response to drug treatment. Chromatin immunoprecipitation sequencing data sets were coupled with transcriptomic results to pinpoint the impact of proscillaridin A on gene pathways associated with specific chromatin modifications. Lastly, we evaluated the effect of the combination of proscillaridin A and the DNA demethylating drug decitabine on eRMS RD cell growth and clonogenic potential.
Results
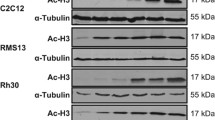
Clinically relevant concentration of proscillaridin A (5 nM) produced growth inhibition, cell cycle arrest and loss of clonogenicity in eRMS RD cells. Proscillaridin A produced a significant downregulation of MYC protein expression and inhibition of oncogenic transcriptional programs controlled by MYC, involved in cell replication. Interestingly, significant reduction in total histone 3 acetylation and on specific lysine residues (lysine 9, 14, 18, and 27 on histone 3) was associated with significant protein downregulation of a series of lysine acetyltransferases (KAT3A, KAT3B, KAT2A, KAT2B, and KAT5). In addition, proscillaridin A produced synergistic growth inhibition and loss of clonogenicity when combined with the approved DNA demethylating drug decitabine.
Conclusion
Proscillaridin A produces anticancer and epigenetic effects in the low nanomolar range and its combination with decitabine warrants further investigation for the treatment of eRMS.






Similar content being viewed by others
References
Dasgupta R, Fuchs J, Rodeberg D (2016) Rhabdomyosarcoma. Semin Pediatr Surg 25(5):276–283
Vella S, Pomella S, Leoncini PP, Colletti M, Conti B, Marquez VE et al (2015) MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin Epigenetics 7:82
van Erp AEM, Versleijen-Jonkers YMH, van der Graaf WTA, Fleuren EDG (2018) Targeted therapy-based combination treatment in rhabdomyosarcoma. Mol Cancer Ther 17(7):1365–1380
Jawad N, McHugh K (2019) The clinical and radiologic features of paediatric rhabdomyosarcoma. Pediatr Radiol 49(11):1516–1523
Gartrell J, Pappo A (2020) Recent advances in understanding and managing pediatric rhabdomyosarcoma. F1000Res 9:(Faculty Rev):685
HaDuong JH, Martin AA, Skapek SX, Mascarenhas L (2015) Sarcomas. Pediatr Clin N Am 62(1):179–200
Williams RF, Fernandez-Pineda I, Gosain A (2016) Pediatric sarcomas. Surg Clin N Am 96(5):1107–1125
Dziuba I, Kurzawa P, Dopierala M, Larque AB, Januszkiewicz-Lewandowska D (2018) Rhabdomyosarcoma in children—current pathologic and molecular classification. Pol J Pathol 69(1):20–32
Pappo AS, Dirksen U (2018) Rhabdomyosarcoma, ewing sarcoma, and other round cell sarcomas. J Clin Oncol 36(2):168–179
Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M et al (2009) RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer 48(11):975–982
Sun X, Guo W, Shen JK, Mankin HJ, Hornicek FJ, Duan Z (2015) Rhabdomyosarcoma: advances in molecular and cellular biology. Sarcoma 2015:232010
Yohe ME, Gryder BE, Shern JF, Song YK, Chou HC, Sindiri S, et al (2019) MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 10(448:eaan4470.).
Dang CV (2012) MYC on the path to cancer. Cell 149(1):22–35
Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al (2010) Selective inhibition of BET bromodomains. Nature 468(7327):1067–1073
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146(6):904–917
Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR (2015) BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 58(6):1028–1039
Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D et al (2015) BET inhibitor resistance emerges from leukaemia stem cells. Nature 525(7570):538–542
Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S et al (2016) Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep 16(5):1273–1286
Cochran AG, Conery AR, Sims RJ 3rd (2019) Bromodomains: a new target class for drug development. Nat Rev Drug Discov 18(8):609–628
Marampon F, Gravina GL, Di Rocco A, Bonfili P, Di Staso M, Fardella C et al (2011) MEK/ERK inhibitor U0126 increases the radiosensitivity of rhabdomyosarcoma cells in vitro and in vivo by downregulating growth and DNA repair signals. Mol Cancer Ther 10(1):159–168
Gravina GL, Festuccia C, Popov VM, Di Rocco A, Colapietro A, Sanita P et al (2016) c-Myc sustains transformed phenotype and promotes radioresistance of embryonal rhabdomyosarcoma cell lines. Radiat Res 185(4):411–422
Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y et al (2015) Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 6:7557
Marchesi I, Fiorentino FP, Rizzolio F, Giordano A, Bagella L (2012) The ablation of EZH2 uncovers its crucial role in rhabdomyosarcoma formation. Cell Cycle 11(20):3828–3836
Raynal NJ, Lee JT, Wang Y, Beaudry A, Madireddi P, Garriga J et al (2016) Targeting calcium signaling induces epigenetic reactivation of tumor suppressor genes in cancer. Can Res 76(6):1494–1505
Raynal NJ, Da Costa EM, Lee JT, Gharibyan V, Ahmed S, Zhang H et al (2017) Repositioning FDA-approved drugs in combination with epigenetic drugs to reprogram colon cancer epigenome. Mol Cancer Ther 16(2):397–407
Da Costa EM, McInnes G, Beaudry A, Raynal NJ (2017) DNA methylation-targeted drugs. Cancer J 23(5):270–276
Jones PA, Issa JP, Baylin S (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17(10):630–641
Da Costa EM, Armaos G, McInnes G, Beaudry A, Moquin-Beaudry G, Bertrand-Lehouillier V et al (2019) Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. J Exp Clin Cancer Res 38(1):251
Prassas I, Diamandis EP (2008) Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov 7(11):926–935
Prassas I, Paliouras M, Datti A, Diamandis EP (2008) High-throughput screening identifies cardiac glycosides as potent inhibitors of human tissue kallikrein expression: implications for cancer therapies. Clin Cancer Res 14(18):5778–5784
Ianevski A, Giri AK, Aittokallio T (2020) SynergyFinder 20: visual analytics of multi-drug combination synergies. Nucleic Acids Res 48(1):488–493
Ianevski A, He L, Aittokallio T, Tang J (2020) SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics 36(8):2645
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102(43):15545–15550
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38(4):576–589
Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM et al (2019) Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 51(12):1714–1722
Stewart E, McEvoy J, Wang H, Chen X, Honnell V, Ocarz M et al (2018) Identification of therapeutic targets in rhabdomyosarcoma through integrated genomic, epigenomic, and proteomic analyses. Cancer Cell 34(3):411–26 e19
Ciarapica R, Carcarino E, Adesso L, De Salvo M, Bracaglia G, Leoncini PP et al (2014) Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer 14:139
Pomella S, Sreenivas P, Gryder BE, Wang L, Milewski D, Cassandri M et al (2021) Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in fusion negative rhabdomyosarcoma. Nat Commun 12(1):192
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ et al (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107(50):21931–21936
Steinberger J, Robert F, Halle M, Williams DE, Cencic R, Sawhney N et al (2019) Tracing MYC expression for small molecule discovery. Cell Chem Biol 26(5):699-710 e6
Gregory MA, Hann SR (2000) c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol 20(7):2423–2435
Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ et al (2004) The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol 24(24):10826–10834
Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E et al (2005) Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol 25(23):10220–10234
Currie GM, Wheat JM, Kiat H (2011) Pharmacokinetic considerations for digoxin in older people. Open Cardiovasc Med J 5:130–135
Funding
This work was supported by the Charles-Bruneau Foundation and Upcycle grant from the Cancer Research Society to N J-M R. N J-M R holds a research scholar Junior 2 award from the Fonds de Recherche du Québec en Santé.
Author information
Authors and Affiliations
Contributions
NJ-MR designed the study. CR, MH, RD, MC and PSt-O, performed experimental studies and bioinformatic analyses. All authors participated in the writing of the manuscript. The datasets generated during the current study are available from the corresponding author on reasonable request.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary figure S1
: Gene expression data (RPKM) from RNA-sequencing of RD cells (untreated and treated with proscillaridin A, 5nM, 48h, n=3) of a series of genes involved in cell proliferation (A) and differentiation (B) in RMS cells. Genes showed a significant difference in gene expression between untreated (Untr.) and treated conditions. Log2 fold changes are indicated in the figure. A letter ‘’a’’ indicates p<0.05 (n=3) (TIF 21 KB)
Supplementary figure S2: Levels of KAT6A (MOZ), KAT7 (HBO1) and histone 4 acetylation after proscillaridin A treatment in RD cells
. (A-B) Measurement of KAT6A and KAT7 expression levels after a time-course of proscillaridin A treatment (5 nM, 8h to 96h) in RD cells. (A) KAT6A (35 kDa) and KAT7 (70 kDa) expression levels were assessed by western blotting in RD cells. ACTIN was used as loading control. (B) KAT6A and KAT7 expression levels were quantified and expressed as a percentage of untreated cells (n=3). (C-D) Measurement of histone 4 acetylation (H4) levels after a time-course proscillaridin A treatment (5 nM, 8h to 96h) in RD cells. (C) H4 acetylation levels were assessed using antibodies against H4K5ac, H4K8ac, H4K16ac, H4K20ac, and total H4 pan-acetylation (PanAc). H4 was used as loading control (8 kDa). (D) H4 acetylation levels were quantified and expressed as a percentage of untreated cells (n=3) (TIF 2595 KB)
Rights and permissions
About this article
Cite this article
Huot, M., Caron, M., Richer, C. et al. Repurposing proscillaridin A in combination with decitabine against embryonal rhabdomyosarcoma RD cells. Cancer Chemother Pharmacol 88, 845–856 (2021). https://doi.org/10.1007/s00280-021-04339-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-021-04339-6