Abstract
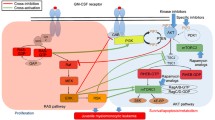
The current study analyzed the clinical and genetic characteristics of a family with familial myeloproliferative neoplasms (MPNs). Whole-exome sequencing was conducted, and a germline heterozygous mutation in lysine methyltransferase 2A (KMT2A, also known as MLL1), G3131S (c.9391G > A, p.Gly3131Ser, rs150804738), was identified. Somatic DNA and germline DNA were collected from 8 family members, 120 healthy donors (somatic DNA), and 30 healthy donors (germline DNA). Using Sanger sequencing, the KMT2A G3131S mutation was analyzed. Four individuals, the proband (II-1), his sister (patient II-2), and family members II-3 and III-1 (somatic DNA and germline DNA), tested positive for the KMT2A G3131S mutation. We did not observe the KMT2A G3131S mutation in healthy donors (somatic DNA and germline DNA), indicating that this is not a SNP. Bioinformatics analysis of KMT2A G3131S suggested that protein structure changes could be caused by this mutation. To further elucidate the function of KMT2A G3131S, the CRISPR-Cas9 technique was applied to generate a KMT2A G3131S heterozygous K562 cell line. The colony formation potency, apoptosis, and cell cycle of KMT2A G3131S mutant K562 cells were analyzed. The results demonstrated that KMT2A G3131S mutant K562 cells showed increased proliferation and colony formation ability. Immunophenotyping was performed using flow cytometry to analyze the surface marker expression of gene-edited KMT2A G3131S mutant K562 cells. A significant increase in CD11b and mild increases in CD61 and CD235a were observed in KMT2A G3131S mutant K562 cells, suggesting that the KMT2A G3131S mutant could cause an increase in myeloproliferation. May-Giemsa staining showed that the morphological changes in KMT2A G3131S mutant K562 cells were consistent with the flow cytometry analysis. To verify which downstream genes were affected by the KMT2A G3131S mutant, we performed real-time PCR to evaluate the expression of previously reported KMT2A-related genes and found that C-MYB expression was significantly decreased. Western blotting was applied to investigate the expression of Kmt2a and C-myb proteins, and the results showed that in KMT2A G3131S mutant K562 cells, the expression of C-myb was decreased. Our findings suggested that KMT2A G3131S could affect the myeloproliferation of K562 cells and decrease C-myb expression. In conclusion, KMT2A G3131S could be considered a novel genetic susceptibility gene in familial MPN.





Similar content being viewed by others
References
Sud A, Chattopadhyay S, Thomsen H, Sundquist K, Sundquist J, Houlston RS, Hemminki K (2018) Familial risks of acute myeloid leukemia, myelodysplastic syndromes, and myeloproliferative neoplasms. Blood 132(9):973–976
Goldin LR, Bjorkholm M, Kristinsson SY, Samuelsson J, Landgren O (2009) Germline and somatic JAK2 mutations and susceptibility to chronic myeloproliferative neoplasms. Genome Med 1(5):55
Bellanne-Chantelot C, Jego P, Lionne-Huyghe P, Tulliez M, Najman A (2008) French group on myeloproliferative d: The JAK2(V617F) mutation may be present several years before the occurrence of overt myeloproliferative disorders. Leukemia 22(2):450–451
Trifa AP, Banescu C, Tevet M, Bojan A, Dima D, Urian L, Torok-Vistai T, Popov VM, Zdrenghea M, Petrov L et al (2016) TERT rs2736100 A>C SNP and JAK2 46/1 haplotype significantly contribute to the occurrence of JAK2 V617F and CALR mutated myeloproliferative neoplasms - a multicentric study on 529 patients. Br J Haematol 174(2):218–226
Belcic Mikic T, Pajic T, Sever M (2019) CALR mutations in a cohort of JAK2 V617F negative patients with suspected myeloproliferative neoplasms. Sci Rep 9(1):19838
Artinger EL, Mishra BP, Zaffuto KM, Li BE, Chung EK, Moore AW, Chen Y, Cheng C, Ernst P (2013) An MLL-dependent network sustains hematopoiesis. Proc Natl Acad Sci U S A 110(29):12000–12005
Meyer C, Burmeister T, Groger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS et al (2018) The MLL recombinome of acute leukemias in 2017. Leukemia 32(2):273–284
Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM (2010) c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest 120(2):593–606
Clarke M, Volpe G, Sheriff L, Walton D, Ward C, Wei W, Dumon S, Garcia P, Frampton J (2017) Transcriptional regulation of SPROUTY2 by MYB influences myeloid cell proliferation and stem cell properties by enhancing responsiveness to IL-3. Leukemia 31(4):957–966
Garcia P, Clarke M, Vegiopoulos A, Berlanga O, Camelo A, Lorvellec M, Frampton J (2009) Reduced c-Myb activity compromises HSCs and leads to a myeloproliferation with a novel stem cell basis. EMBO J 28(10):1492–1504
Landgren O, Goldin LR, Kristinsson SY, Helgadottir EA, Samuelsson J, Bjorkholm M (2008) Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24,577 first-degree relatives of 11,039 patients with myeloproliferative neoplasms in Sweden. Blood 112(6):2199–2204
Mora B, Rumi E, Guglielmelli P, Barraco D, Maffioli M, Rambaldi A, Caramella M, Komrokji R, Gotlib J, Kiladjian JJ et al (2019) Second primary malignancies in postpolycythemia vera and postessential thrombocythemia myelofibrosis: A study on 2233 patients. Cancer Med 8(9):4089–4092
Rumi E, Passamonti F, Della Porta MG, Elena C, Arcaini L, Vanelli L, Del Curto C, Pietra D, Boveri E, Pascutto C et al (2007) Familial chronic myeloproliferative disorders: clinical phenotype and evidence of disease anticipation. J Clin Oncol 25(35):5630–5635
Bellanne-Chantelot C, Chaumarel I, Labopin M, Bellanger F, Barbu V, De Toma C, Delhommeau F, Casadevall N, Vainchenker W, Thomas G et al (2006) Genetic and clinical implications of the Val617Phe JAK2 mutation in 72 families with myeloproliferative disorders. Blood 108(1):346–352
Malak S, Labopin M, Saint-Martin C, Bellanne-Chantelot C, Najman A (2012) French Group of Familial Myeloproliferative D: Long term follow up of 93 families with myeloproliferative neoplasms: life expectancy and implications of JAK2V617F in the occurrence of complications. Blood Cells Mol Dis 49(3–4):170–176
McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ (2007) Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1(3):338–345
Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378(6556):505–508
Winters AC, Bernt KM (2017) MLL-rearranged leukemias-an update on science and clinical approaches. Front Pediatr 5:4
Tkachuk DC, Kohler S, Cleary ML (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71(4):691–700
Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E (1992) The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71(4):701–708
Zeleznik-Le NJ, Harden AM, Rowley JD (1994) 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci U S A 91(22):10610–10614
Yano T, Nakamura T, Blechman J, Sorio C, Dang CV, Geiger B, Canaani E (1997) Nuclear punctate distribution of ALL-1 is conferred by distinct elements at the N terminus of the protein. Proc Natl Acad Sci U S A 94(14):7286–7291
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 24(1):88–91
Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK (2002) The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 30(4):958–965
Chen J, Santillan DA, Koonce M, Wei W, Luo R, Thirman MJ, Zeleznik-Le NJ, Diaz MO (2008) Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res 68(15):6199–6207
Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10(5):1107–1117
Richmond J, Carol H, Evans K, High L, Mendomo A, Robbins A, Meyer C, Venn NC, Marschalek R, Henderson M et al (2015) Effective targeting of the P53-MDM2 axis in preclinical models of infant MLL-rearranged acute lymphoblastic leukemia. Clin Cancer Res 21(6):1395–1405
Al-Kershi S, Bhayadia R, Ng M, Verboon L, Emmrich S, Gack L, Schwarzer A, Strowig T, Heckl D, Klusmann JH (2019) The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia. Blood Adv 3(24):4252–4263
Somers K, Kosciolek A, Bongers A, El-Ayoubi A, Karsa M, Mayoh C, Wadham C, Middlemiss S, Neznanov N, Kees UR et al (2020) Potent antileukemic activity of curaxin CBL0137 against MLL-rearranged leukemia. Int J Cancer 146(7):1902–1916
Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T (2008) Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 22(8):1539–1541
Tan YT, Sun Y, Zhu SH, Ye L, Zhao CJ, Zhao WL, Chen Z, Chen SJ, Liu H (2016) Deregulation of HOX genes by DNMT3A and MLL mutations converges on BMI1. Leukemia 30(7):1609–1612
Schwieger M, Schuler A, Forster M, Engelmann A, Arnold MA, Delwel R, Valk PJ, Lohler J, Slany RK, Olson EN et al (2009) Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. Blood 114(12):2476–2488
Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, Jiang X, Hong GM, Kunjamma RB, Ren H et al (2013) PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood 121(8):1422–1431
Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H et al (2016) PBX3 and MEIS1 cooperate in hematopoietic cells to drive acute myeloid leukemias characterized by a core transcriptome of the MLL-rearranged disease. Cancer Res 76(3):619–629
Funding
This work was supported in part by the National Natural Science Foundation of China (Nos. 82070175, 81800198, 81700168) and the Natural Science Foundation of Hunan Province (No. 2019JJ50863).
Author information
Authors and Affiliations
Contributions
Professor Hongling Peng diagnosed patient and designed and composed this study. Dr. Zhao Cheng designed and composed this study and wrote the manuscript. Miss Le Yin and Sisi Xie collected data and conducted the study. Mr. Yi Chen helped with performing experiments. Mr. Wang Li and Xian Jiang helped with data collection and experiments. Dr. Heng Li and Dr. Ji Li helped with clinic sample collection. Mr. Zefang Wu helped with clinic sample collection. Dr. Xiang Xiao helped with flow cytometry and RNA reverse-transcription and transcriptome sequencing. Professor Guangsen Zhang partially participate the study and helped with clinic sample collection.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Second Xiangya Hospital, Central South University. Informed consent was obtained in writing in accordance with the Declaration of Helsinki.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yin, L., Xie, S., Chen, Y. et al. Novel germline mutation KMT2A G3131S confers genetic susceptibility to familial myeloproliferative neoplasms. Ann Hematol 100, 2229–2240 (2021). https://doi.org/10.1007/s00277-021-04562-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-021-04562-4