Abstract
Purpose
Positron emission tomography (PET) using 18F-3′-deoxy-3′-fluorothymidine ([18F]FLT) allows noninvasive monitoring of tumour proliferation. For serial imaging in individual patients, good reproducibility is essential. The purpose of the present study was to evaluate the reproducibility of quantitative [18F]FLT measurements.
Methods
Nine patients with non-small-cell lung cancer (NSCLC) and six with head-and-neck cancer (HNC) underwent [18F]FLT PET twice within 7 days prior to therapy. The maximum pixel value (SUVmax) and a threshold defined volume (SUV41%) were defined for all delineated lesions. The plasma to tumour transfer constant (Ki) was estimated using both Patlak graphical analysis and nonlinear regression (NLR). NLR was also used to estimate k3, which, at least in theory, selectively reflects thymidine kinase 1 activity. The level of agreement between test and retest values was assessed using the intraclass correlation coefficient (ICC) and Bland-Altman analysis.
Results
All primary tumours and >90% of clinically suspected locoregional metastases could be delineated. In total, 24 lesions were defined. NLR-derived Ki, Patlak-derived Ki, SUV41% and SUVmax showed excellent reproducibility with ICCs of 0.92, 0.95, 0.98 and 0.93, and SDs of 16%, 12%, 7% and 11%, respectively. Reproducibility was poor for k3 with an ICC of 0.43 and SD of 38%.
Conclusion
Quantitative [18F]FLT measurements are reproducible in both NSCLC and HNC patients. When monitoring response in individual patients, changes of more than 15% in SUV41%, 20–25% in SUVmax and Patlak-derived Ki, and 32% in NLR3k-derived Ki are likely to represent treatment effects.
Similar content being viewed by others
Introduction
18F-3′-Deoxy-3′-fluorothymidine ([18F]FLT) has been proposed as a positron emission tomography (PET) tracer of proliferation. FLT is a substrate for thymidine kinase 1 (TK1), which is a key enzyme in the salvage pathway of thymidine DNA synthesis. Several studies have shown good correlations between [18F]FLT uptake and other markers of cellular proliferation, including proliferating cell nuclear antigen, flow cytometry and Ki-67 nuclear staining [1–5].
In most tumours [18F]FLT uptake is lower than [18F]FDG uptake [6]. In addition, its biodistribution and metabolic profile are not ideal [7]. Consequently, [18F]FDG PET remains the method of choice for diagnosing and staging of tumours. It has been suggested that [18F]FLT may have additional value in combination with [18F]FDG because the combination has been reported to result in fewer false-positive findings [8, 9], but results have not been consistent [2, 10, 11].
Based on its TK1-related signal, [18F]FLT PET has been proposed as a biomarker for predicting (early) response to systemic or locoregional treatment [12–14]. Recent studies have shown that [18F]FLT PET can predict response to therapy as early as 1 week after treatment [15], and a decrease in [18F]FLT uptake seems to be correlated with prolonged overall survival [16, 17].
In addition, [18F]FLT might also be a specific tracer for monitoring the effects of agents targeting thymidylate synthase (TS), an enzyme that plays a central role in the de novo pathway of DNA synthesis. Downregulation of this pathway results in an upregulation of the salvage pathway and thus of [18F]FLT uptake. Recent results have indicated that anti-TS effects can be monitored as soon as 2 hours after administration [18].
To evaluate response in individual patients, reproducibility of the parameter of interest needs to be known. The purpose of the present study was to determine reproducibility of quantitative [18F]FLT measurements in a prospective study of patients with non-small-cell lung cancer (NSCLC) or head-and-neck cancer (HNC).
Materials and methods
Eligible patients were included prospectively after providing written informed consent in accordance with institutional review board approval. In total, nine patients with NSCLC and six with HNC were scanned twice within 7 days (mean 1.9 days, median 1 day) prior to any therapy using an ECAT EXACT HR+ scanner (Siemens/CTI, Knoxville, TN). This scanner has an axial field of view (FOV) of 15 cm, divided into 63 contiguous planes. Each patient was positioned supine on the scanner bed with the primary tumour in the centre of the axial FOV. The primary tumour and all other evaluable lesions in the FOV were analysed. A venous catheter was placed in all patients, which was used for injection of [18F]FLT and venous blood sampling [19]. In addition, in HNC patients an arterial catheter was placed in the radial artery for arterial blood sampling. Each acquisition started with a 10 to 15-min transmission scan in order to acquire a fixed number of 85 million counts [20], which was used for attenuation correction of the subsequent emission scan. This was followed by a bolus injection of 370 MBq [18F]FLT in 5 ml saline through an injector (Medrad International, Maastricht, The Netherlands) at 0.8 ml/s, after which the line was flushed with 42 ml saline (2.0 ml/s). Simultaneously with the injection of [18F]FLT, a dynamic emission scan (in 2-D acquisition mode) was started with a total duration of 60 min and with variable frame lengths (6×5 s, 6×10 s, 3×20 s, 5×30 s, 5×60 s, 8×150 s, and 6×300 s). All dynamic scan data were corrected for dead time, decay, scatter, randoms and photon attenuation, and were reconstructed as 128×128 matrices using filtered back projection (FBP) with a Hanning filter (cut-off, 0.5 cycles per pixel). This resulted in a transaxial spatial resolution of around 7 mm full-width at half-maximum (FWHM). As FBP reconstructed images suffer from streak artefacts, volume of interest (VOI) definition may be inaccurate, especially in lesions with low tumour-to-background contrast [21, 22]. Therefore, for VOI definition purposes only, the last three frames of the sinograms (45–60 min after injection) were summed and reconstructed using ordered-subset expectation maximization (OSEM) with two iterations and 16 subsets followed by postsmoothing of the reconstructed images using a 5-mm FWHM gaussian filter to obtain the same resolution as for the FBP images [21].
In the HNC patients, arterial 18F concentrations were monitored continuously using a fully automated blood sampling device (Veenstra Instruments, Joure, The Netherlands) [23]. The withdrawal rate was 5 ml/min during the first 10 min and 2.5 ml/min thereafter. In all patients, venous blood samples were drawn at 5, 10, 20, 30, 40 and 60 min after injection to correct for plasma/whole blood ratios and plasma metabolite fractions, and as a quality control procedure for defining an image-derived input function (IDIF) for NSCLC patients as described for [18F]FDG [24]. To avoid contamination, 3–5 ml blood was withdrawn prior to each sample and the line was flushed with 1.5 ml saline after sampling, as described previously [19].
Venous blood samples were analysed using solid-phase extraction chromatography for separation of [18F]FLT from [18F]FLT glucuronide. For this procedure 0.3 ml plasma was dissolved in 2 ml water. This solution was placed onto a SepPak Vac 6cc (1 g) C18 cartridge (Waters Corporation, Milford, MA). The eluate was collected, after which the cartridge was rinsed with 5 ml water to collect the polar metabolites, being primarily [18F]FLT-glucuronide. The cartridge was then rinsed with 5 ml 96% ethanol to collect the parent compound. All fractions and the cartridge were counted using a Wallac 1480 Wizard well counter (Perkin-Elmer Life Science, Zaventem, Belgium), and the percentage parent within each plasma sample calculated. Metabolite data were interpolated by fitting to an exponential function.
Image analysis
The maximum pixel value within the tumour and a threshold defined volume (41% of the maximum pixel value with correction for local background) were defined semiautomatically for any lesion with adequate focal uptake [25, 26]. Boundaries of lesions without adequate tumour-to-background contrast (low focal uptake and/or high level of background) are difficult to define and, consequently, delineation is hampered by increased observer variation. Therefore, only lesions which required no or only minor manual delineation were included. In the latter case background values were set to zero for voxels directly adjacent to the VOI that had a physiologically high radiotracer concentration (hypervascular areas, haematopoietic bone marrow and liver). After this procedure the threshold technique was applied, resulting in volumes specifically containing tumour. Tumour VOIs were defined on OSEM reconstructed images and transferred to FBP reconstructed dynamic images, thereby generating time–activity curves (TACs). An input function was obtained by continuous arterial blood sampling in HNC patients and derived from the dynamic FBP images in NSCLC patients. IDIFs were defined by multiple manually drawn 2-D regions of interest (ROIs) over the aortic arch (elliptical ROI, 8×30 mm) and ascending aorta (spherical ROI, 15 mm) [27]. Again, ROIs were then projected onto all frames to generate input TACs.
Data analysis
Full kinetic analysis to derive values of the individual rate constants and Ki, the plasma to tumour transfer constant, was performed for threshold defined VOIs using the following methods: (1) Patlak graphical analysis (time interval 10–60 min after injection; Ki only) [28] and (2) nonlinear regression (NLR) using the standard two-tissue compartment model with both three (NLR3k) and four (NLR4k) rate constants and an additional blood volume parameter (Vb). In both cases, the metabolite-corrected plasma curve was used as input function. The presence of a fourth rate constant (representing dephosphorylation of phosphorylated [18F]FLT) and the need to include this in the model were assessed by comparing fits with and without a k4 parameter using Akaike [29] and Schwarz [30] criteria.
Kinetic analysis of regional tissue TACs was performed using dedicated software developed within Matlab (The Mathworks, Natick, MA). In general, fits are rejected when the (fitted) standard error of any parameter exceeds 25%. This was, however, never the case in the present study. In addition, the goodness of fit was checked visually for all TACs. SUV was derived for both the maximum pixel value (SUVmax) and the threshold defined VOI (SUV41%) for the time intervals 40–60 min and 50–60 min after injection (with multiple correction factors [31]). The level of agreement between test and retest values was assessed using the intraclass correlation coefficient (ICC) with a two-way random model with absolute agreement, and Bland-Altman analysis [32]. In the latter case, the percentage difference in values (Δ) between two measures was plotted against (1) the mean of both measures and (2) the mean threshold defined volume. In this way, possible dependency on both absolute [18F]FLT uptake and tumour size could be evaluated. Additionally, dependency was statistically analysed using linear regression. Finally, a one-sample t test was applied to the Δ values to assess systematic bias and the parameter values of both test and retest scans were compared using the Wilcoxon signed ranks test to evaluate the distribution.
Results
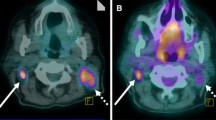
All primary tumours and more than 90% of clinically suspected locoregional metastases could be delineated. In total, 24 lesions (15 NSCLC and 9 HNC) were defined. A small subset of lesions (5/24) required manual intervention during delineation (Fig. 1). Full kinetic analysis was possible for 23 lesions; blood analyses failed in one patient. The median lesion size (threshold defined PET volume) was 8.2 cm3 (range 1.7–86.1 cm3), the median NLR3k-derived Ki was 0.047 ml·min−1·ml−1 (range 0.021–0.120 ml·min−1·ml−1), the median Patlak-derived Ki was 0.037 ml·min−1·ml−1 (range 0.017–0.074 ml·min-1·ml-1), the median SUV41% was 3.3 (range 1.3–6.4) and the median SUVmax was 4.8 (range 2.4–9.3). The 3k model provided better fits than the 4k model in 26 (57%) and 28 (61%) of 46 measured lesions according to Akaike and Schwarz criteria, respectively. In other words, the majority of the data were best fitted with the 3k model. Visual check of the TACs revealed good fits for all lesions.
Descriptive statistics for all parameters of [18F]FLT kinetics for both test and retest scans, as well as p values for the Wilcoxon signed ranks test, are presented in Table 1. No systematic bias (one-sample t test: SUV41%, p=0.98; Patlak Ki, p=0.25), or a significant difference in the distribution of paired differences (Wilcoxon signed ranks test; p>0.08 for all) was found. The threshold VOI definition technique resulted in consistent volumes for most tumours throughout both scans. The median change in tumour volume between test and retest scans was 10%. Only the delineation of one lesion resulted in an exceptional difference in volume of 150% between the two scans. This was probably due to inaccurate definition of the maximum pixel value, possibly caused by patient motion in one or both scans.
Only very small variations were found between different SUV41% normalization methods (body weight, body surface area and lean body mass) and time intervals (40–60 min and 50–60 min after injection) with ICCs ranging from 0.97 to 0.98. Therefore, the SUV results are presented for only one correction method (body weight) and a single time interval (40–60 min after injection). NLR3k-derived Ki, Patlak-derived Ki, SUV41% and SUVmax showed excellent reproducibility with ICCs of 0.92, 0.95, 0.98 and 0.93, and SDs of 16%, 12%, 7% and 11%, respectively (Table 2). In contrast, NLR3k-derived k3, which, at least in theory, selectively reflects TK1 activity, showed poor reproducibility with an ICC of 0.43 and an SD of 38%. As expected from Akaike and Schwarz analyses, inclusion of a fourth rate constant resulted in decreased reproducibility with an ICC of 0.75 and an SD of 25% for NLR4k-derived Ki, and an ICC of 0.33 and an SD of 70% for NLR4k-derived k3 (Table 2).
These data show that, when monitoring therapy effects, changes of less than 32% in NLR3k-derived Ki, 24% in Patlak-derived Ki, 14% in SUV41% and 22% in SUVmax (<1.96×SD) cannot be distinguished from normal test–retest variability.
Subgroup analysis for NSCLC and HNC patients showed similar results for Patlak-derived Ki and an even better SD result for NLR3k-derived Ki in NSCLC patients as compared to HNC patients, suggesting no error due to the use of an IDIF (Table 2).
Bland-Altman plots of SUV41% and Patlak-derived Ki showed no dependency on absolute [18F]FLT uptake, but a trend of reduced reproducibility for smaller lesions (Fig. 2). This trend, however, was not statistically significant (SUV41%, p=0.35; Patlak Ki, p=0.51).
Bland-Altman plots for SUV41% and Patlak Ki (circles NSCLC lesions, triangles NHC lesions, delta values percentage change between test and retest scans, SUV41% and SUVmax for the time interval 40–60 min after injection and corrected for body weight, Patlak Patlak-derived Ki, mean PET volume mean threshold defined volume of the test and retest scan in cm3, straight lines mean values, dotted lines ±1.96×SD)
Discussion
Our results show that quantitative [18F]FLT measurements are reproducible in both NSCLC and HNC patients. When serial measurements in patients are performed, changes of more than 15% in SUV41%, 20–25% in Patlak-derived Ki and SUVmax, and 30–35% in NLR3k-derived Ki are likely to represent biological effects. In addition, no significant dependency between absolute [18F]FLT uptake and lesion size was found, implying that the same threshold can be used for all tumour lesions. Although one tumour lesion showed a change of 150% in tumour volume between the two scans, the threshold technique resulted in reproducible tumour volumes for all other lesions.
Akaike and Schwarz analyses showed that for most tumour lesions data were best fitted with the 3k model, indicating that [18F]FLT phophorylation was irreversible, at least within the time frame of the measurements. Therefore, the fourth rate constant should be set to zero. In a recent study it was shown that significant dephosphorylation starts at ±50 min after injection and that k4 can reliably be estimated with a scan time of 120 min [33]. Furthermore, a strong correlation was found between NLR3k Ki derived from a 60-min scan and NLR4k Ki derived from a 120-min scan [33]. A scan protocol of 120 min, however, is too long for routine (serial) clinical studies, especially in critically ill patients. In addition, the risk of movement artefacts increases with longer scan times. Our results indicate that with a scan time of 60 min, a fourth rate constant can be neglected.
To the best of our knowledge, reproducibility of [18F]FLT measurements in malignant tumours has only been assessed in eight patients (nine lesions) with breast cancer [15] and in six patients with NSCLC [34]. Shields et al. [34] evaluated an unknown number of lesions in six patients using SUVmax and SUVmean (50% threshold technique) and carried out full kinetic analyses (NLR with unknown number of rate constants and Patlak) with the use of an IDIF. Unfortunately, full details of their study (e.g. the number of rate constants in the NLR analyses, location and number of lesions, whether mean or median for absolute difference of test and retest scans) were not reported. Nevertheless, reproducibility results were comparable with the present results (around 20% test–retest variability in SUV and kinetic parameters).
Results in breast cancer [15] are also consistent with the present results with reported ICCs of 0.99 and 0.97, and SDs of 11% and 15% for SUV and Patlak Ki, respectively. Differences in study design and methods, however, make it difficult to perform a direct comparison. The absence of IDIF analyses, the relatively low number of lesions included with no locoregional (lymph node) metastases, and the very high tumour-to-background ratio in the breast (due to low uptake of [18F]FLT in fatty tissue) [35, 36] imply that results for breast tumours do not necessarily hold for tumours of other origins (with lower tumour-to-background ratio).
In the present study IDIF was used for NSCLC lesions, thereby greatly simplifying the acquisition protocol for thoracic tumours. Clearly, this is much more convenient for patients and better suited for routine clinical use. Subgroup analyses for NLR3k and Patlak-derived Ki did not reveal inferior results for NSCLC as compared to HNC, where arterial blood sampling was performed, suggesting that the use of an IDIF does not significantly contribute to test–retest variability.
In the present analyses, locoregional lymph nodes were also included, and thereby all possible scenarios encountered in clinical trials were evaluated. In addition, this provided a means for evaluating possible dependency on absolute [18F]FLT value and tumour size.
SUVmax is the most frequently used (semi)quantitative measure in clinical PET studies due to its ease of definition and low observer variation. Nevertheless, results indicate that better reproducibility can be achieved by using threshold defined SUV41%. Although pulmonary lesions are not easy to delineate due to physiologically high uptake in surrounding mediastinum, liver, vertebrae and costal bones, reproducibility was not reduced, indicating that the applied threshold technique is reliable.
Since radiotracer uptake depends on perfusion and extraction, SUV and Ki are not specific measurements of TK1 activity. [18F]FLT uptake mainly depends on the extracellular ATP concentration, which has greatest impact on the turnover of [18F]FLT due to induction of a change in structure of TK1 from a dimer to a tetramer, which has about a 20-fold greater effect on the turnover of the tracer [37]. However, previous data have shown that [18F]FLT uptake may also depend on perfusion (blood flow) rather than on TK1 activity [38]. Therefore, at least in theory, k3 appears to be the parameter of interest for studying TK1 activity. Unfortunately, NLR3k-derived k3 showed poor reproducibility. However, both Ki and SUV have shown good correlations with other proliferation markers, with Ki having the strongest correlation [4, 15, 33]. The present results indicate that both Ki and SUV are reproducible.
Since Ki has the best correlation with other proliferation markers, dynamic scanning should, whenever possible, be applied in response monitoring studies. If static scans are acquired, however, the present results support the use of a threshold defined VOI instead of the maximum pixel value. In the present study only therapy-naive tumours were analysed. It should be kept in mind, however, that an intervention may affect the various rate constants in a different manner. For example, a certain therapy could result in a reduction in delivery (K1) with only a minor change in TK1 activity (k3). In that case, both Ki and SUV would decrease more than the actual decrease in proliferation rate. In addition, SUV and Ki might not be similarly affected.
Conclusion
Our results show that both simplified and full kinetic analyses of [18F]FLT data have excellent reproducibility in NSCLC and HNC patients. Furthermore, the data support the use of an IDIF for kinetic analyses of intrathoracic lesions. Future response monitoring studies, correlating [18F]FLT response with pathological and clinical outcome, should be performed to assess which [18F]FLT parameter is best for predicting response to therapy.
References
Yamamoto Y, Nishiyama Y, Ishikawa S, Nakano J, Chang SS, Bandoh S, et al. Correlation of 18F-FLT and 18F-FDG uptake on PET with Ki-67 immunohistochemistry in non-small cell lung cancer. Eur J Nucl Med Mol Imaging 2007;34:1610–6.
Troost EG, Vogel WV, Merkx MA, Slootweg PJ, Marres HA, Peeters WJ, et al. 18F-FLT PET does not discriminate between reactive and metastatic lymph nodes in primary head and neck cancer patients. J Nucl Med 2007;48:726–35.
Kenny LM, Vigushin DM, Al-Nahhas A, Osman S, Luthra SK, Shousha S, et al. Quantification of cellular proliferation in tumor and normal tissues of patients with breast cancer by [18F]fluorothymidine-positron emission tomography imaging: evaluation of analytical methods. Cancer Res 2005;65:10104–12.
Vesselle H, Grierson J, Muzi M, Pugsley JM, Schmidt RA, Rabinowitz P, et al. In vivo validation of 3′deoxy-3′-[(18)F]fluorothymidine ([(18)F]FLT) as a proliferation imaging tracer in humans: correlation of [(18)F]FLT uptake by positron emission tomography with Ki-67 immunohistochemistry and flow cytometry in human lung tumors. Clin Cancer Res 2002;8:3315–23.
Barthel H, Cleij MC, Collingridge DR, Hutchinson OC, Osman S, He Q, et al. 3′-deoxy-3′-[18F]fluorothymidine as a new marker for monitoring tumor response to antiproliferative therapy in vivo with positron emission tomography. Cancer Res 2003;63:3791–8.
Salskov A, Tammisetti VS, Grierson J, Vesselle H. FLT: measuring tumor cell proliferation in vivo with positron emission tomography and 3′-deoxy-3′-[18F]fluorothymidine. Semin Nucl Med 2007;37:429–39.
Dittmann H, Dohmen BM, Paulsen F, Eichhorn K, Eschmann SM, Horger M, et al. [18F]FLT PET for diagnosis and staging of thoracic tumours. Eur J Nucl Med Mol Imaging 2003;30:1407–12.
Yamamoto Y, Nishiyama Y, Kimura N, Ishikawa S, Okuda M, Bandoh S, et al. Comparison of (18)F-FLT PET and (18)F-FDG PET for preoperative staging in non-small cell lung cancer. Eur J Nucl Med Mol Imaging 2008;35:236–45.
Buck AK, Halter G, Schirrmeister H, Kotzerke J, Wurziger I, Glatting G, et al. Imaging proliferation in lung tumors with PET: 18F-FLT versus 18F-FDG. J Nucl Med 2003;44:1426–31.
Cobben DC, Elsinga PH, Hoekstra HJ, Suurmeijer AJ, Vaalburg W, Maas B, et al. Is 18F-3′-fluoro-3′-deoxy-L-thymidine useful for the staging and restaging of non-small cell lung cancer? J Nucl Med 2004;45:1677–82.
Shields AF. PET imaging with 18F-FLT and thymidine analogs: promise and pitfalls. J Nucl Med 2003;44:1432–4.
Buck AK, Kratochwil C, Glatting G, Juweid M, Bommer M, Tepsic D, et al. Early assessment of therapy response in malignant lymphoma with the thymidine analogue [18F]FLT. Eur J Nucl Med Mol Imaging 2007;34:1775–82.
Herrmann K, Wieder HA, Buck AK, Schöffel M, Krause BJ, Fend F, et al. Early response assessment using 3′-deoxy-3′-[18F]fluorothymidine-positron emission tomography in high-grade non-Hodgkin’s lymphoma. Clin Cancer Res 2007;13:3552–8.
Apisarnthanarax S, Alauddin MM, Mourtada F, Ariga H, Raju U, Mawlawi O, et al. Early detection of chemoradioresponse in esophageal carcinoma by 3′-deoxy-3′-3H-fluorothymidine using preclinical tumor models. Clin Cancer Res 2006;12:4590–7.
Kenny L, Coombes RC, Vigushin DM, Al-Nahhas A, Shousha S, Aboagye EO. Imaging early changes in proliferation at 1 week post chemotherapy: a pilot study in breast cancer patients with 3′-deoxy-3′-[18F]fluorothymidine positron emission tomography. Eur J Nucl Med Mol Imaging 2007;34:1339–47.
Chen W, Delaloye S, Silverman DH, Geist C, Czernin J, Sayre J, et al. Predicting treatment response of malignant gliomas to bevacizumab and irinotecan by imaging proliferation with [18F] fluorothymidine positron emission tomography: a pilot study. J Clin Oncol 2007;25:4714–21.
Kasper B, Egerer G, Gronkowski M, Haufe S, Lehnert T, Eisenhut M, et al. Functional diagnosis of residual lymphomas after radiochemotherapy with positron emission tomography comparing FDG- and FLT-PET. Leuk Lymphoma 2007;48:746–53.
Perumal M, Pillai RG, Barthel H, Leyton J, Latigo JR, Forster M, et al. Redistribution of nucleoside transporters to the cell membrane provides a novel approach for imaging thymidylate synthase inhibition by positron emission tomography. Cancer Res 2006;66:8558–64.
Hoekstra CJ, Hoekstra OS, Lammertsma AA. On the use of the injection catheter for venous blood sampling in quantitative FDG PET studies. Eur J Nucl Med 2000;27:1579.
Boellaard R, van Lingen A, van Balen SCM, Lammertsma AA. Optimization of attenuation correction for positron emission tomography studies of thorax and pelvis using count-based transmission scans. Phys Med Biol 2004;49:N31–8.
Boellaard R, van Lingen A, Lammertsma AA. Experimental and clinical evaluation of iterative reconstruction (OSEM) in dynamic PET: quantitative characteristics and effects on kinetic modeling. J Nucl Med 2001;42:808–17.
Riddell C, Carson RE, Carrasquillo JA, Libutti SK, Danforth DN, Whatley M, et al. Noise reduction in oncology FDG PET images by iterative reconstruction: a quantitative assessment. J Nucl Med 2001;42:1316–23.
Boellaard R, van Lingen A, van Balen SC, Hoving BG, Lammertsma AA. Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. Eur J Nucl Med 2001;28:81–9.
Hoekstra CJ, Hoekstra OS, Lammertsma AA. On the use of image-derived input functions in oncological fluorine-18 fluorodeoxyglucose positron emission tomography studies. Eur J Nucl Med 1999;26:1489–92.
Boellaard R, Oyen WJ, Hoekstra CJ, Hoekstra OS, Visser EP, Willemsen AT, et al. The Netherlands protocol for standardisation and quantification of FDG whole body PET studies in multi-centre trials. Eur J Nucl Med Mol Imaging 2008; [Epub ahead of print]. doi:10.1007/s00259-008-0954-3
Erdi YE, Mawlawi O, Larson SM, Imbriaco M, Yeung H, Finn R, et al. Segmentation of lung lesion volume by adaptive positron emission tomography image thresholding. Cancer 1997;80:2505–9.
van der Weerdt AP, Klein LJ, Boellaard R, Visser CA, Visser FC, Lammertsma AA. Image-derived input functions for determination of MRGlu in cardiac (18)F-FDG PET scans. J Nucl Med 2001;42:1622–9.
Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 1983;3:1–7.
Akaike H. A new look at the statistical identification. IEEE Trans Automat Contr 1978;19:716–23.
Schwarz G. Estimating the dimension of a model. Ann Statist 1978;6:461–4.
Hoekstra CJ, Paglianiti I, Hoekstra OS, Smit EF, Postmus PE, Teule GJ, et al. Monitoring response to therapy in cancer using [18F]-2-fluoro-2-deoxy-D-glucose and positron emission tomography: an overview of different analytical methods. Eur J Nucl Med 2000;27:731–43.
Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1:307–10.
Muzi M, Vesselle H, Grierson JR, Mankoff DA, Schmidt RA, Peterson L, et al. Kinetic analysis of 3′-deoxy-3′-fluorothymidine PET studies: validation studies in patients with lung cancer. J Nucl Med 2005;46:274–82.
Shields AF, Lawhorn-Crews JM, Briston DA, Douglas KA, Mangner TJ, Muzik O. The reproducibility of FLT PET in patients with untreated non-small cell lung cancer. J Nucl Med 2005;46:426P.
Smyczek-Gargya B, Fersis N, Dittmann H, Vogel U, Reischl G, Machulla HJ, et al. PET with [18F]fluorothymidine for imaging of primary breast cancer: a pilot study. Eur J Nucl Med Mol Imaging 2004;31:720–4.
Been LB, Elsinga PH, de Vries J, Cobben DC, Jager PL, Hoekstra HJ, et al. Positron emission tomography in patients with breast cancer using (18)F-3′-deoxy-3′-fluoro-l-thymidine ((18)F-FLT) – a pilot study. Eur J Surg Oncol 2006;32:39–43.
Dimitrakopoulou-Strauss A, Strauss LG. The role of 18F-FLT in cancer imaging: does it really reflect proliferation? Eur J Nucl Med Mol Imaging 2008;35:523–6.
Muzi M, Spence AM, O’Sullivan F, Mankoff DA, Wells JM, Grierson JR, et al. Kinetic analysis of 3′-deoxy-3′-18F-fluorothymidine in patients with gliomas. J Nucl Med 2006;47:1612–21.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Langen, A.J., Klabbers, B., Lubberink, M. et al. Reproducibility of quantitative 18F-3′-deoxy-3′-fluorothymidine measurements using positron emission tomography. Eur J Nucl Med Mol Imaging 36, 389–395 (2009). https://doi.org/10.1007/s00259-008-0960-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-008-0960-5