Abstract
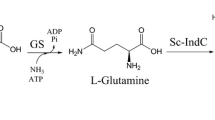
Indigoidine is a dark-blue natural pigment with application prospect and synthesized from glutamine (Gln) by series of indigoidine synthetases (IndCs). Indigoidine production can be improved by enhancing Gln pool via supplementing Gln directly or converting metabolism glutamate (Glu) to Gln by glutamine synthetase (GlnA). But, Gln is expensive, and excess Gln inhibits indigoidine production of the recombinant strain. Supplementing Glu instead of Gln may improve the productive and economic efficiency of indigoidine, but the local activities and positions of the indigoidine pathway enzymes GlnA, Sc-IndC, and the helper protein of Sc-IndC (IndB) should be well arranged. We identified the Streptomyces chromofuscus ATCC 49982 derived IndC (Sc-IndC) as an more efficient IndC compared to other IndCs applied for constructing indigoidine-producting strains, and designed series of protein scaffold complexes with architectures of PDZ, SH3, and GBD domains (PxSyG1) to arrange the pathway enzymes. The strain recruiting GlnA, Sc-IndC, and IndB on the PDZ, SH3, and GBD domains of scaffold P1S2G1, respectively, was the most efficient. In the strain, the GlnA supplied sufficient local Gln for Sc-IndC from Glu, and the generated Gln was immediately consumed by Sc-IndC to relieve cell growth inhibition caused by Gln. The optimum Glu concentration (6 g/L) for the strain was higher than those of the strains recruiting Sc-IndC on the GBD domain, which was away from the PDZ domain recruiting GlnA. The highest titer of indigoidine was 12 g/L, which was two folds of the control without scaffold (5.8 g/L). The titer is 5 g/L higher than the control without Glu supplemented (6.9 g/L), meaning that 97% of the supplemented Glu was transformed into indigoidine. The batch fermentation with the optimum strain in a 5-L reactor achieved an indigoidine titer of 14 g/L in 60 h. To our knowledge, this was the most efficient indigoidine productivity achieved so far. The optimization strategies by protein scaffold should be applicative to other pathways with complex substrate demands.
Key points
•Protein scaffold systems were designed to arrange the indigoidine synthetic pathway.
•The scaffold system improved supplement of Gln for indigoidine production from Glu.
•The inhibition caused by excess Gln was relieved by proper designed scaffold.
•The yield and titer of indigoidine was improved by arranging the pathway enzymes.

Graphical abstract





Similar content being viewed by others
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Brachmann AO, Kirchner F, Kegler C, Kinski SC, Schmitt I, Bode HB (2012) Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157(1):96–99
Cude WN, Mooney J, Tavanaei AA, Hadden MK, Frank AM, Gulvik CA, May AL, Buchan A (2012) Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine roseobacter Phaeobacter sp. strain Y4I. Appl Environ Microb 78(14):4771–4780
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27(8):753–759. https://doi.org/10.1038/nbt.1557
Feng G, Zhu M, Liu L, Li C (2019) A quantitative one-pot synthesis method for industrial azo pigments with recyclable wastewater. Green Chem 21(7):1769–1776. https://doi.org/10.1039/c8gc03982h
Flassig RJ, Fachet M, Hoffner K, Barton PI, Sundmacher K (2016) Dynamic flux balance modeling to increase the production of high-value compounds in green microalgae. Biotechnol Biofuels 9:12. https://doi.org/10.1186/s13068-016-0556-4
Galanopoulou AP, Moraїs S, Georgoulis A, Morag E, Bayer EA, Hatzinikolaou DG (2016) Insights into the functionality and stability of designer cellulosomes at elevated temperatures. Appl Microbiol Biotechnol 100(20):8731–8743
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343–345. https://doi.org/10.1038/nmeth.1318
Graser TA, Godel HG, Albers S, F?Ldi P, Fürst P (1985) An ultra rapid and sensitive high-performance liquid chromatographic method for determination of tissue and plasma free amino acids. Anal Biochem 151(1):142–152
Gromek SM, Suria AM, Fullmer MS, Garcia JL, Peter GJ, Nyholm SV, Balunas MJ (2016) Leisingera sp. JC1, a bacterial isolate from Hawaiian bobtail squid eggs, produces indigoidine and differentially inhibits vibrios. Front Microbiol 7. https://doi.org/10.3389/fmicb.2016.01342
Han GH, Bang SE, Babu BK, Chang M, Shin HJ, Si WK (2010) Bio-indigo production in two different fermentation systems using recombinant Escherichia coli cells harboring a flavin-containing monooxygenase gene ( fmo). Process Biochem 150(3):369–369
Kim H, Siu KH, Raeeszadeh-Sarmazdeh M, Sun Q, Chen Q, Chen W (2015) Bioengineering strategies to generate artificial protein complexes. Biotechnol Bioeng 112(8):1495–1505. https://doi.org/10.1002/bit.25637
Kim HJ, Jang S, Kim J, Yang YH, Kim YG, Kim BG, Choi KY (2017) Biosynthesis of indigo in Escherichia coli expressing self-sufficient CYP102A from Streptomyces cattleya. Dyes Pigments 140:29–35. https://doi.org/10.1016/j.dyepig.2017.01.029
Kong L, Xu G, Liu X, Wang J, Tang Z, Cai Y-S, Shen K, Tao W, Zheng Y, Deng Z, Price NPJ, Chen W (2019) Divergent biosynthesis of C-nucleoside ninimycin and indigoidine in bacteria. iScience 22:430–440. https://doi.org/10.1016/j.isci.2019.11.037
Kuhlborn J, Danner A-K, Frey H, Iyer R, Arduengo AJ III, Opatz T (2017) Examples of xylochemistry: colorants and polymers. Green Chem 19(16):3780–3786. https://doi.org/10.1039/c7gc01244f
Li PW, Li J, Guo ZY, Tang W, Han JS, Meng XX, Hao TT, Zhu YX, Zhang LX, Chen YH (2015) An efficient blue-white screening based gene inactivation system for Streptomyces. Appl Microbiol Biotechnol 99(4):1923–1933
Liu QP, Xie NN, He Y, Wang L, Shao YC, Zhao HZ, Chen FS (2014) MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Appl Microbiol Biotechnol 98(1):285–296
Moon TS, Dueber JE, Shiue E, Prather KL (2010) Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng 12(3):298–305. https://doi.org/10.1016/j.ymben.2010.01.003
Namgung S, Park HA, Kim J, Lee P-G, Kim B-G, Yang Y-H, Choi K-Y (2019) Ecofriendly one-pot biosynthesis of indigo derivative dyes using CYP102G4 and PrnA halogenase. Dyes Pigments 162:80–88. https://doi.org/10.1016/j.dyepig.2018.10.009
Owen JG, Calcott MJ, Robins KJ, Ackerley DF (2016) Generating functional recombinant NRPS enzymes in the laboratory setting via peptidyl carrier protein engineering. Cell Chem Biol 23(11):1–12
Pearson JT, Dabrowski MJ, Kung I, Atkins WM (2005) The central loop of Escherichia coli glutamine synthetase is flexible and functionally passive. Arch Biochem Biophys 436(2):397–405
Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C (2001) Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291(5509):1790–1792
Price JV, Chen L, Whitaker WB, Papoutsakis E, Chen W (2016) Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc Natl Acad Sci U S A 113(45):12691–12696. https://doi.org/10.1073/pnas.1601797113
Reverchon S, Nasser W, Robert-Baudouy J (2010) pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol 11(6):1127–1139
Satagopan S, Sun Y, Parquette JR, Tabita FR (2017) Synthetic CO2-fixation enzyme cascades immobilized on self-assembled nanostructures that enhance CO2/O-2 selectivity of RubisCO. Biotechnol Biofuels 10. https://doi.org/10.1186/s13068-017-0861-6
Takahashi H, Kumagai T, Kitani K, Mori M, Matoba Y, Sugiyama M (2007) Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J Biol Chem 282(12):9073–9081. https://doi.org/10.1074/jbc.M611319200
Vaitekunas J, Gasparaviciute R, Rutkiene R, Tauraite D, Meskys R (2016) A 2-hydroxypyridine catabolism pathway in Rhodococcus rhodochrous strain PY11. Appl Environ Microb 82(4):1264–1273. https://doi.org/10.1128/aem.02975-15
Wehrs M, Prahl J-P, Moon J, Li Y, Tanjore D, Keasling JD, Pray T, Mukhopadhyay A (2018) Production efficiency of the bacterial non-ribosomal peptide indigoidine relies on the respiratory metabolic state in S. cerevisiae. Microb Cell Fact 17:193
Wehrs M, Gladden JM, Liu Y, Platz L, Prahl J-P, Moon J, Papa G, Sundstrom E, Geiselman GM, Tanjore D, Keasling JD, Pray TR, Simmons BA, Mukhopadhyay A (2019) Sustainable bioproduction of the blue pigment indigoidine: expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides. Green Chem 21(12):3394–3406. https://doi.org/10.1039/c9gc00920e
Xia JP, Sheng WH, Su X, Yue FM, Ping ZR, ShuYing H, Kai L, Hui XX (2012) Pathway redesign for deoxyviolacein biosynthesis in Citrobacter freundii and characterization of this pigment. Appl Microbiol Biotechnol 94(6):1521–1532
Xu FC, Gage D, Zhan JX (2015) Efficient production of indigoidine in Escherichia coli. J Ind Microbiol Biotechnol 42(8):1149–1155. https://doi.org/10.1007/s10295-015-1642-5
Yu DY, Xu FC, Valiente J, Wang SY, Zhan JX (2013) An indigoidine biosynthetic gene cluster from Streptomyces chromofuscus ATCC 49982 contains an unusual IndB homologue. J Ind Microbiol Biotechnol 40(1):159–168. https://doi.org/10.1007/s10295-012-1207-9
Yumusak C, Prochazkova AJ, Apaydin DH, Seelajaroen H, Sariciftci NS, Weiter M, Krajcovic J, Qin Y, Zhang W, Zhan J, Kovalenko A (2019) Indigoidine – biosynthesized organic semiconductor. Dyes Pigments 171. https://doi.org/10.1016/j.dyepig.2019.107768
Acknowledgments
We are indebted to Dr. John E Dueber at University of California, Berkeley for providing plasmids for scaffold protein expression.
Funding
This work was supported by grants from the National Natural Science Foundation of China (21708003, 31470787) and Department of Science and Technology of Jilin Province (20190902014TC, 20170519015JH).
Author information
Authors and Affiliations
Contributions
LW and DY conceived and designed the experiments. LW, YS, DL, BL, and YG performed the experiments. LW, YS, DL, and DY analyzed data. LW and DY wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 471 kb)
Rights and permissions
About this article
Cite this article
Wang, L., Sun, Y., Lv, D. et al. Protein scaffold optimizes arrangement of constituent enzymes in indigoidine synthetic pathway to improve the pigment production. Appl Microbiol Biotechnol 104, 10493–10502 (2020). https://doi.org/10.1007/s00253-020-10990-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10990-5