Abstract
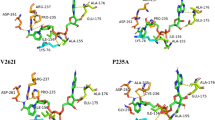
Methyl parathion hydrolase (MPH) that hydrolyzes a wide range of organophosphorus pesticides can be used to remediate land polluted by the pesticides. Here, the catalytic efficiency of methyl parathion hydrolase from Pseudomonas sp. (WBC-3) was enhanced by searching and engineering a critical site far away from the binding pocket. In the first round, a four-site mutant with a modest increased catalytic efficiency (3.2-fold kcat/Km value of the wild type) was obtained with random mutagenesis. By splitting and re-combining the four substitutions in the mutant, the critical site S277, was identified to show the most significant effects of improving binding affinity and catalytic efficiency. With further site-saturation mutagenesis focused on the residue S277, another two substitutions were discovered to have even more significant decrease in Km (40.2 and 47.6 μM) and increased in kcat/Km values (9.5- and 10.3-fold of the wild type) compared to the original four-site mutant (3.0- and 3.2-fold). In the three-dimensional structure, residue S277 is located at a hinge region of a loop, which could act as a “lid” at the substrate entering to the binding pocket. This suggests that substitutions of residue S277 could affect substrate binding via conformational change in substrate entrance region. This work provides a valuable protocol combining random mutagenesis, site-saturation mutagenesis, structural and bioinformatics analyses to obtain mutants with high catalytic efficiency from a screening library of a modest size (3200 strains).




Similar content being viewed by others
References
Abou-Donia MB (2003) Organophosphorus ester-induced chronic neurotoxicity. Arch Environ Health 58(8):484–497
Aktar MW, Sengupta D, Chowdhury A (2009) Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol 2(1):1–12
Alkotaini B, Han NS, Kim BS (2016) Enhanced catalytic efficiency of endo-β-agarase I by fusion of carbohydrate-binding modules for agar prehydrolysis. Enzym Microb Technol 93–94:142–149
Balci H, Ozturk MT, Pijning T, Ozturk SI, Gumusel F (2014) Improved activity and pH stability of E. coli ATCC 11105 penicillin acylase by error-prone PCR. Appl Microbiol Biotechnol 98(10):4467–4477
Benning M, Kuo J, Raushel F, Holden H (1995) Three-dimensional structure of the binuclear metal center of phosphotriesterase. Biochem 34(25):7973–7978
Chen Y, Zhang X, Liu H, Wang Y, Xia X (2002) Study on Pseudomonas sp. WBC-3 capable of complete degradation of methyl parathion. Wei Sheng Wu Xue Bao 42(4):490–497
Cui Z, Li S, Fu G (2001) Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl Environ Microbiol 67(10):4922–4925
Dong YJ, Bartlam M, Sun L, Zhou YF, Zhang ZP, Zhang CG, Rao Z, Zhang XE (2005) Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3. J Mol Biol 353(3):655–663
Ekkhunnatham A, Jongsareejit B, Yamkunthong W, Wichitwechkarn J (2012) Purification and characterization of methyl parathion hydrolase from Burkholderia cepacia capable of degrading organophosphate insecticides. World J Microbiol Biotechnol 28(4):1739–1746
Ely F, Foo JL, Jackson CJ, Gahan LR, Ollis DL, Schenk G (2007) Enzymatic bioremediation: organophosphate degradation by binuclear metallo-hydrolases. Curr Top Biochem Res 9:63–78
Engqvist MK, McIsaac RS, Dollinger P, Flytzanis NC, Abrams M, Schor S, Arnold FH (2015) Directed evolution of Gloeobacter violaceus rhodopsin spectral properties. J Mol Biol 427(1):205–220
Fan Y, Fang W, Xiao Y, Yang X, Zhang Y, Bidochka MJ, Yan P (2007) Directed evolution for increased chitinase activity. Appl Microbiol Biotechnol 76(1):135–139
Fu G, Cui Z, Huang T, Li S (2004) Expression, purification, and characterization of a novel methyl parathion hydrolase. Protein Expr Purif 36(2):170–176
Ghanem E, Raushel FM (2005) Detoxification of organophosphate nerve agents by bacterial phosphotriesterase. Toxicol Appl Pharmacol 207(2):459–470
Gong Y, Xu GC, Chen Q, Yin JG, Li CX, Xu JH (2016) Iterative multitarget evolution dramatically enhances the enantioselectivity and catalytic efficiency of Bacillus subtilis esterase towards bulky benzoate esters of DL-menthol. Catal Sci Technol 6(7):2370–2376
Hovmoller S, Zhou T, Ohlson T (2002) Conformations of amino acids in proteins. Acta Crystallogr Sect D: Biol Crystallogr 58(5):768–776
Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53(1):315–351
Jang CH, Piao YL, Huang X, Yoon EJ, Park SH, Lee K, Zhan CG, Cho H (2016) Modeling and re-engineering of Azotobacter vinelandii alginate lyase to enhance its catalytic efficiency for accelerating biofilm degradation. PLoS One 11(6):e0156197
Jensen AF, Petersen A, Granby K (2003) Cumulative risk assessment of the intake of organophosphorus and carbamate pesticides in the Danish diet. Food Addit Contam 20(8):776–785
Jeong YS, Choi JM, Kyeong HH, Choi JY, Kim EJ, Kim HS (2014) Rational design of organophosphorus hydrolase with high catalytic efficiency for detoxifying a V-type nerve agent. Biochem Biophys Res Commun 449(3):263–267
Josse D, Xie W, Renault F, Rochu D, Schopfer LM, Masson P, Lockridge O (1999) Identification of residues essential for human paraoxonase (PON1) arylesterase/organophosphatase activities. Biochem 38(9):2816–2825
Kou CL, La Du BN (1998) Calcium binding by human and rabbit serum paraoxonases-structural stability and enzymatic activity. Drug Metab Dispos 26(7):653–660
Li X, He J, Li S (2007) Isolation of a chlorpyrifos-degrading bacterium, Sphingomonas sp. strain Dsp-2, and cloning of the mpd gene. Res Microbiol 158(2):143–149
Li Z, Huang M, Gu Z, Holler TP, Cheng L, Hong Y, Li C (2016) Asp577 mutations enhance the catalytic efficiency of cyclodextrin glycosyltransferase from Bacillus circulans. Int J Biol Macromol 83:111–116
Li W, Xu S, Zhang B, Zhu Y, Hua Y, Kong X, Sun L, Hong J (2017) Directed evolution to improve the catalytic efficiency of urate oxidase from Bacillus subtilis. PLoS One 12(5):e0177877
Luo XJ, Zhao J, Li CX, Bai YP, Reetz MT, Yu HL, Xu JH (2016) Combinatorial evolution of phosphotriesterase toward a robust malathion degrader by hierarchical iteration mutagenesis. Biotechnol Bioeng 113(11):2350–2357
Molloy S, Nikodinovic Runic J, Martin LB, Hartmann H, Solano F, Decker H, O’Connor KE (2013) Engineering of a bacterial tyrosinase for improved catalytic efficiency towards D-tyrosine using random and site directed mutagenesis approaches. Biotechnol Bioeng 110(7):1849–1857
Ng TK, Gahan LR, Schenk G, Ollis DL (2015) Altering the substrate specificity of methyl parathion hydrolase with directed evolution. Arch Biochem Biophys 573:59–68
Qayyum H, Maroof H, Yasha K (2009) Remediation and treatment of organopollutants mediated by peroxidases: a review. Crit Rev Biotechnol 29(2):94–119
Scott C, Pandey G, Hartley CJ, Jackson CJ, Cheesman MJ, Taylor MC, Pandey R, Khurana JL, Teese M, Coppin CW (2008) The enzymatic basis for pesticide bioremediation. Indian J Microbiol 48(1):65–79
Trollope KM, Görgens JF, Volschenk H (2015) Semirational directed evolution of loop regions in Aspergillus japonicus β-fructofuranosidase for improved fructooligosaccharide production. Appl Environ Microbiol 81(20):7319–7329
Watkins LM, Mahoney HJ, Mcculloch JK, Raushel FM (1997) Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase. J Biol Chem 272(41):25596–25601
Yang J, Yang C, Jiang H, Qiao C (2008) Overexpression of methyl parathion hydrolase and its application in detoxification of organophosphates. Biodegradation 19(6):831–839
Zou SP, Zheng YG, Wu Q, Wang ZC, Xue YP, Liu ZQ (2018) Enhanced catalytic efficiency and enantioselectivity of epoxide hydrolase from Agrobacterium radiobacter AD1 by iterative saturation mutagenesis for (R)-epichlorohydrin synthesis. Appl Microbiol Biotechnol 102(2):733–742
Funding
This work was supported by the 1000 Plan of China (K2069999) and NSFC of Jiangsu Province (BK20151126), NSFC (51603089) to FX, National Natural Science Foundation of China (21406089) to HQY.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 709 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Yang, H. & Xu, F. Identifying and engineering a critical amino acid residue to enhance the catalytic efficiency of Pseudomonas sp. methyl parathion hydrolase. Appl Microbiol Biotechnol 102, 6537–6545 (2018). https://doi.org/10.1007/s00253-018-9108-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9108-0