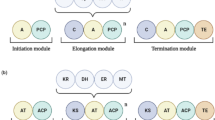
Abstract
Neomycin, an aminoglycoside antibiotic, is widely used in the livestock husbandry due to its higher antimicrobial activity and availability of feed additives in animals. However, its production yield is relatively low and cannot meet the needs of developing market and clinical application. Here, the entire natural neo cluster was cloned from Streptomyces fradiae CGMCC 4.576 by φBT1 integrase-mediated site-specific recombination. Then, the rational reconstruction of the neo cluster was performed by using λ-Red-mediated PCR targeting for improving neomycin production. In order to coordinate with this attempt, the supplementation of suitable precursors was carried out. The constructed recombinant strain Sf/pKCZ03 has multi-copy of the neo cluster modified by disrupting the negative regulatory gene neoI and replacing the native promoter of the neoE-D with PkasO*. Compared to the yield (1282 mg/L) of Streptomyces fradiae CGMCC 4.576, the engineered strain Sf/pKCZ03 had a 36% enhancement of neomycin production. Quantitative real-time PCR analysis revealed the increased transcription of structural genes (neoE, neoB, neoL, aacC8) and regulatory genes (neoR, neoH) in Sf/pKCZ03. Additionally, under the supplementation of 1 g/L N-acetyl-D-glucosamine and 5 g/L L-glutamine, the yield of engineered strain Sf/pKCZ03 showed 62% and 107% improvements compared to that of the wild-type strain in the original medium, respectively. These findings demonstrated that engineering the antibiotic gene cluster in combination with precursors feeding was an effective approach for strain improvement, and would be potentially extended to other Streptomyces for large-scale production of commercialized antibiotics.






Similar content being viewed by others
References
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. https://doi.org/10.1038/417141a
Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49
Bottero V, Sadagopan S, Johnson KE, Dutta S, Veettil MV, Chandran B (2013) Kaposi’s sarcoma-associated herpesvirus-positive primary effusion lymphoma tumor formation in NOD/SCID mice is inhibited by neomycin and neamine blocking angiogenin’s nuclear translocation. J Virol 87(21):11806–11820. https://doi.org/10.1128/jvi.01920-13
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–6645. https://doi.org/10.1073/pnas.120163297
Du D, Wang L, Tian Y, Liu H, Tan H, Niu G (2015) Genome engineering and direct cloning of antibiotic gene clusters via phage phi BT1 integrase-mediated site-specific recombination in Streptomyces. Sci Rep 5:8740. https://doi.org/10.1038/srep08740
Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF, Zhang Y (2012) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446. https://doi.org/10.1038/nbt.2183
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100(4):1541–1546
Huang F, Li Y, Yu J, Spencer JB (2002) Biosynthesis of aminoglycoside antibiotics: cloning, expression and characterisation of an aminotransferase involved in the pathway to 2-deoxystreptamine. Chem Commun (Camb) 23:2860–2861
Huang F, Spiteller D, Koorbanally NA, Li Y, Llewellyn NM, Spencer JB (2007) Elaboration of neosamine rings in the biosynthesis of neomycin and butirosin. Chembiochem 8(3):283–288
Jacela JY, Derouchey JM, Tokach MD, Goodband RD, Nelssen JL, Renter DG, Dritz SS (2009) Feed additives for swine: fact sheets - acidifiers and antibiotics. J Swine Health Prod 17(5):270–275
Jiang W, Zhu T (2016) Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments. Nat Protoc 11:960–975. https://doi.org/10.1038/nprot.2016.055
Jiang L, Wei J, Li L, Niu G, Tan H (2013) Combined gene cluster engineering and precursor feeding to improve gougerotin production in Streptomyces graminearus. Appl Microbiol Biotechnol 97(24):10469–10477. https://doi.org/10.1007/s00253-013-5270-6
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Kudo F, Eguchi T (2009) Biosynthetic enzymes for the aminoglycosides butirosin and neomycin. Methods Enzymol 459:493–519. https://doi.org/10.1016/S0076-6879(09)04620-5
Kudo F, Yamamoto Y, Yokoyama K, Eguchi T, Kakinuma K (2005) Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. J Antibiot (Tokyo) 58(12):766–774
Kudo F, Fujii T, Kinoshita S, Eguchi T (2007) Unique O-ribosylation in the biosynthesis of butirosin. Bioorg Med Chem 15(13):4360–4368
Kudo F, Hoshi S, Kawashima T, Kamachi T, Eguchi T (2014) Characterization of a radical S-adenosyl-L-methionine epimerase, NeoN, in the last step of neomycin B biosynthesis. J Am Chem Soc 136(39):13909–13915. https://doi.org/10.1021/ja507759f
Kvitko BH, Mcmillan IA, Schweizer HP (2013) An improved method for oriT-directed cloning and functionalization of large bacterial genomic regions. Appl Environ Microbiol 79(16):4869–4878. https://doi.org/10.1128/AEM.00994-13
Lee NC, Larionov V, Kouprina N (2015) Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res 43(8):e55–e55. https://doi.org/10.1093/nar/gkv112
Li Y, Tan H (2017) Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci China Life Sci 60(9):935–938. https://doi.org/10.1007/s11427-017-9115-x
Li Y, Li J, Tian Z, Xu Y, Zhang J, Liu W, Tan H (2016) Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J Biol Chem 291(10):5406–5417. https://doi.org/10.1074/jbc.M115.695874
Li L, Zheng G, Chen J, Ge M, Jiang W, Lu Y (2017) Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab Eng 40:80–92. https://doi.org/10.1016/j.ymben.2017.01.004
Liu Y, Wu Y, Zhang X, Hu G, Wu Y (2016) Neamine inhibits growth of pancreatic cancer cells in vitro and in vivo. J Huazhong Univ Sci Technol Med Sci 36(1):82–87. https://doi.org/10.1007/s11596-016-1546-2
Liu P, Zhu H, Zheng G, Jiang W, Lu Y (2017) Metabolic engineering of Streptomyces coelicolor for enhanced prodigiosins (RED) production. Sci China Life Sci 60(9):948–957. https://doi.org/10.1007/s11427-017-9117-x
Liu M, Li S, Xie Y, Jia S, Hou Y, Zou Y, Zhong C (2018) Enhanced bacterial cellulose production by Gluconacetobacter xylinus via expression of Vitreoscilla hemoglobin and oxygen tension regulation. Appl Microbiol Biotechnol 102(3):1155–1165. https://doi.org/10.1007/s00253-017-8680-z
Lu C, Zhang X, Jiang M, Bai L (2016) Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng 35:129–137. https://doi.org/10.1016/j.ymben.2016.02.012
Meng X, Wang W, Xie Z, Li P, Li Y, Guo Z, Lu Y, Yang J, Guan K, Lu Z, Tan H, Chen Y (2017) Neomycin biosynthesis is regulated positively by AfsA-g and NeoR in Streptomyces fradiae CGMCC 4.7387. Sci China Life Sci 60(9):980–991. https://doi.org/10.1007/s11427-017-9120-8
Niu G, Zheng J, Tan H (2017) Biosynthesis and combinatorial biosynthesis of antifungal nucleoside antibiotics. Sci China Life Sci 60(9):939–947. https://doi.org/10.1007/s11427-017-9116-0
Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ (1999) Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J Bacteriol 181(1):204–211
Pullan ST, Chandra G, Bibb MJ, Merrick M (2011) Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12(1):175. https://doi.org/10.1186/1471-2164-12-175
Pyeon HR, Nah HJ, Kang SH, Choi SS, Kim ES (2017) Heterologous expression of pikromycin biosynthetic gene cluster using Streptomyces artificial chromosome system. Microb Cell Factories 16(1):96. https://doi.org/10.1186/s12934-017-0708-7
Rokem JS, Lantz AE, Nielsen J (2007) Systems biology of antibiotic production by microorganisms. Nat Prod Rep 24(6):1262–1287
Sambrock J, Russel D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Takahashi Y, Igarashi M (2017) Destination of aminoglycoside antibiotics in the ‘post-antibiotic era’. J Antibiot (Tokyo) 71:4–14. https://doi.org/10.1038/ja.2017.117
Tamegai H, Nango E, Koike-Takeshita A, Kudo F, Kakinuma K (2002) Significance of the 20-kDa subunit of heterodimeric 2-deoxy-scyllo-inosose synthase for the biosynthesis of butirosin antibiotics in Bacillus circulans. Biosci Biotechnol Biochem 66(7):1538–1545
Tan G, Peng Y, Lu C, Bai L, Zhong J (2015) Engineering validamycin production by tandem deletion of gamma-butyrolactone receptor genes in Streptomyces hygroscopicus 5008. Metab Eng 28:74–81. https://doi.org/10.1016/j.ymben.2014.12.003
Thanapipatsiri A, Claesen J, Gomezescribano JP, Bibb M, Thamchaipenet A (2015) A Streptomyces coelicolor host for the heterologous expression of type III polyketide synthase genes. Microb Cell Factories 14(1):145. https://doi.org/10.1186/s12934-015-0335-0
Waksman SA, Lechevalier HA (1949) Neomycin, a new antibiotic active against streptomycin-resistant bacteria, including tuberculosis organisms. Science 109(2830):305–307. https://doi.org/10.1126/science.109.2830.305
Wang H, Li Z, Jia R, Hou Y, Yin J, Bian X, Li A, Müller R, Stewart AF, Fu J, Zhang Y (2016) RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat Protoc 11:1175–1190. https://doi.org/10.1038/nprot.2016.054
Wang X, Tian X, Wu Y, Shen X, Yang S, Chen S (2018) Enhanced doxorubicin production by Streptomyces peucetius using a combination of classical strain mutation and medium optimization. Prep Biochem Biotechnol 48:1–8. https://doi.org/10.1080/10826068.2018.1466156
Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30(8):1180–1182
Xu D, Liu G, Lin C, Lu X, Chen W, Deng Z (2013) Identification of Mur34 as the novel negative regulator responsible for the biosynthesis of muraymycin in Streptomyces sp. NRRL30471. PLoS One 8(10):e76068. https://doi.org/10.1371/journal.pone.0076068
Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS (2014) Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A 111(5):1957–1962. https://doi.org/10.1073/pnas.1319584111
Yin J, Hoffmann M, Bian X, Tu Q, Yan F, Xia L, Ding X, Stewart AF, Muller R, Fu J (2015) Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM 41398 in Streptomyces coelicolor A3(2). Sci Rep 5:15081. https://doi.org/10.1038/srep15081
Yokoyama K, Kudo F, Kuwahara M, Inomata K, Tamegai H, Eguchi T, Kakinuma K (2005) Stereochemical recognition of doubly functional aminotransferase in 2-deoxystreptamine biosynthesis. J Am Chem Soc 127(16):5869–5874
Yokoyama K, Ohmori D, Kudo F, Eguchi T (2008a) Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. Biochemistry 47(34):8950–8960. https://doi.org/10.1021/bi800509x
Yokoyama K, Yamamoto Y, Kudo F, Eguchi T (2008b) Involvement of two distinct N-acetylglucosaminyltransferases and a dual-function deacetylase in neomycin biosynthesis. Chembiochem 9(6):865–869. https://doi.org/10.1002/cbic.200700717
Yuan L, Wei H, Feng H, Li S (2006) Rapid analysis of native neomycin components on a portable capillary electrophoresis system with potential gradient detection. Anal Bioanal Chem 385(8):1575–1579
Zeng H, Wen S, Xu W, He Z, Zhai G, Liu Y, Deng Z, Sun Y (2015) Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA (sm) combined system. Appl Microbiol Biotechnol 99(24):10575–10585. https://doi.org/10.1007/s00253-015-6931-4
Zhang Y, Pan G, Zou Z, Fan K, Yang K, Tan H (2013) JadR*-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol Microbiol 90(4):884–897. https://doi.org/10.1111/mmi.12406
Zhu Z, Li H, Yu P, Guo Y, Luo S, Chen Z, Mao X, Guan W, Li Y (2017) SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl Microbiol Biotechnol 101(4):1547–1557. https://doi.org/10.1007/s00253-016-7918-5
Zhuo J, Ma B, Xu J, Hu W, Zhang J, Tan H, Tian Y (2017) Reconstruction of a hybrid nucleoside antibiotic gene cluster based on scarless modification of large DNA fragments. Sci China Life Sci 60(9):968–979. https://doi.org/10.1007/s11427-017-9119-1
Zou Z, Du D, Zhang Y, Zhang J, Niu G, Tan H (2014) A-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol Microbiol 94(3):490–505. https://doi.org/10.1111/mmi.12752
Acknowledgments
We are grateful to Professor Yuhui Sun (School of Pharmaceutical Sciences, Wuhan University, China) for kindly providing the plasmid pWHU2653.
Funding
This work was supported by Beijing Natural Science Foundation (Grant No. 5184034) and the National Natural Science Foundation of China (Grant Nos. 31771378, 31800029, and 31470206).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approved
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 548 kb)
Rights and permissions
About this article
Cite this article
Zheng, J., Li, Y., Guan, H. et al. Enhancement of neomycin production by engineering the entire biosynthetic gene cluster and feeding key precursors in Streptomyces fradiae CGMCC 4.576. Appl Microbiol Biotechnol 103, 2263–2275 (2019). https://doi.org/10.1007/s00253-018-09597-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-09597-8