Abstract
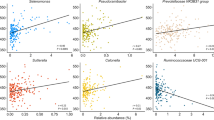
The microbiota of whole crop corn silage and feces of silage-fed dairy cows were examined. A total of 18 dairy cow feces were collected from six farms in Japan and China, and high-throughput Illumina sequencing of the V4 hypervariable region of 16S rRNA genes was performed. Lactobacillaceae were dominant in all silages, followed by Acetobacteraceae, Bacillaceae, and Enterobacteriaceae. In feces, the predominant families were Ruminococcaceae, Bacteroidaceae, Clostridiaceae, Lachnospiraceae, Rikenellaceae, and Paraprevotellaceae. Therefore, Lactobacillaceae of corn silage appeared to be eliminated in the gastrointestinal tract. Although fecal microbiota composition was similar in most samples, relative abundances of several families, such as Ruminococcaceae, Christensenellaceae, Turicibacteraceae, and Succinivibrionaceae, varied between farms and countries. In addition to the geographical location, differences in feeding management between total mixed ration feeding and separate feeding appeared to be involved in the variations. Moreover, a cow-to-cow variation for concentrate-associated families was demonstrated at the same farm; two cows showed high abundance of Succinivibrionaceae and Prevotellaceae, whereas another had a high abundance of Porphyromonadaceae. There was a negative correlation between forage-associated Ruminococcaceae and concentrate-associated Succinivibrionaceae and Prevotellaceae in 18 feces samples. Succinivibrionaceae, Prevotellaceae, p-2534-18B5, and Spirochaetaceae were regarded as highly variable taxa in this study. These findings help to improve our understanding of variation and similarity of the fecal microbiota of dairy cows with regard to individuals, farms, and countries. Microbiota of naturally fermented corn silage had no influence on the fecal microbiota of dairy cows.

Similar content being viewed by others
References
Anderson CL, Schneider CJ, Erickson GE, MacDonald JC, Fernando SC (2016) Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs. J Appl Microbiol 120:588–599. doi:10.1111/jam.13039
Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetenda EG, Wang J, Guarner F, Pedersen O, de Vos DM, Brunak S, Dore J, MetaHIT Consortium, Weissenbach J, Ehrlich SD, Bork P (2011) Enterotypes of the human gut microbiome. Nature 473:174–180. doi:10.1038/nature09944
Beauchemin KA, Wang WZ (2005) Effects of physically effective fiber on intake, chewing activity, and ruminal acidosis for dairy cows fed diets based on corn silage. J Dairy Sci 88:2117–2129. doi:10.3168/jds.S0022-0302(05)72888-5
Benchaar C, Petit HV, Berthiaume R, Ouellet DR, Chiquette J, Chouinard PY (2007) Effects of essential oils on digestion, ruminal fermentation, rumen microbial populations, milk production, and milk composition in dairy cows fed alfalfa silage or corn silage. J Dairy Sci 90:886–897. doi:10.3168/jds.S0022-0302(07)71572-2
Broderick GA, Radloff WJ (2004) Effect of molasses supplementation on the production of lactating dairy cows fed diets based on alfalfa and corn silage. J Dairy Sci 87:2997–3009. doi:10.3168/jds.S0022-0302(04)73431-1
Castillo AR, Kebreab E, Beever DE, Barbi JH, Sutton JD, Kirby HC, France J (2001) The effect of protein supplementation on nitrogen utilization in lactating dairy cows fed grass silage diets. J Anim Sci 79:247–253. doi:10.2527/2001.791247x
Claesson MJ, Wang Q, O’Sullivan O, Diniz RG, Cole JR, Ross RP, O’Toole PW (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200. doi:10.1093/nar/gkq873
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci 107:14691–14706. doi:10.1073/pnas.1005963107
Derakhshani H, De Buck J, Mortier R, HW Barkema, Krause D O, Khafipou E (2016) The features of fecal and ileal mucosa-associated microbiota in dairy calves during early infection with Mycobacterium avium subspecies paratuberculosis. Front Microbiol 7:1–13. doi: 10.3389/fmicb.2016.00426
Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE (2014) Human genetics shape the gut microbiome. Cell 159:789–799. doi:10.1016/j.cell.2014.09.053
Han K, Bose S, Wang J, Kim BS, Kim MJ, Kim EJ, Kim H (2015) Contrasting effects of fresh and fermented kimchi consumption on gut microbiota composition and gene expression related to metabolic syndrome in obese Korean women. Mol Nutr Food Res 59:1004–1008. doi:10.1002/mnfr.201400780
Jami E, Mizrahi I (2012a) Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:1–8. doi:10.1371/journal.pone.0033306
Jami E, Mizrahi I (2012b) Similarity of the ruminal bacteria across individual lactating cows. Anaerobe 18:338–343. doi:10.1016/j.anaerobe.2012.04.003
Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G (2015) Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi:10.1128/AEM.00720-15
Jung JY, Lee SH, Jeon CO (2014) Kimchi microflora: history, current status, and perspectives for industrial kimchi production. Appl Microbiol Biotechnol 98:2385–2393. doi:10.1007/s00253-014-5513-1
Khafipour E, Li S, Tun HM, Derakhshani H, Moossavi S, Plaizier JC (2016) Effects of grain feeding on microbiota in the digestive tract of cattle. Anim Front 6:13–19. doi:10.2527/af.2016-0018
Kim M, Kim J, Kuehn LA, Bono JL, Berry ED, Kalchayanand N, Freetly HC, Benson AK, Wells JE (2014) Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci 92:683–694. doi:10.2527/jas2013-6841
Kraut-Cohen J, Tripathi V, Chen Y, Gatica J, Volchinski V, Sela S, Weinberg Z, Cytryn E (2016) Temporal and spatial assessment of microbial communities in commercial silages from bunker silos. Appl Microbiol Biotechnol 100:6827–6835. doi:10.1007/s00253-016-7512-x
Kurokawa K, Itoh T, Kuwahara T, Oshima K, ToH TA, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M (2007) Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14:169–181. doi:10.1093/dnares/dsm018
Lee KE, Choi UH, Ji GE (1996) Effect of kimchi intake on the composition of human large intestinal bacteria. Korean J Food Sci Technol 28:981–986
Lima FS, Oikonomou G, Lima SF, Bicalho MLS, Ganda EK, Filho JCO, Lorenzo G, Trojacanec P, Bicalhoa RC (2015) Prepartum and postpartum rumen fluid microbiomes: characterization and correlation with production traits in dairy cows. Appl Environ Microbiol 81:1327–1337. doi:10.1128/AEM.03138-14
Liu J, Zhang M, Xue C, Zhu W, Mao S (2016) Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J Dairy Sci 99:1–14. doi:10.3168/jds.2016-11398
Maekawa M, Beauchemin KA, Christensen DA (2002) Effect of concentrate level and feeding management on chewing activities, saliva production, and ruminal pH of lactating dairy cows. J Dairy Sci 85:1165–1175. doi:10.3168/jds.S0022-0302(02)74179-9
Mao S, Zhang M, Liu J, Zhu W (2015) Characterizing the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci Rep 5:16116. doi:10.1038/srep16116
Menezes A, Lewis E, O’Donovan M, O’Neill BF, Clipson N, Doyle EM (2011) Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol Ecol 78:256–265. doi:10.1111/j.1574-6941.2011.01151.x
Ni K, Minh TT, Tu TTM, Tsuruta T, Pang H, Nishino N (2017) Comparative microbiota assessment of wilted Italian ryegrass, whole crop corn, and wilted alfalfa silage using denaturing gradient gel electrophoresis and next-generation sequencing. Appl Microbiol Biotechnol 101:1385–1394. doi:10.1007/s00253-016-7900-2
Nocek JE, Steele RL, Braund DG (1986) Performance of dairy cows fed forage and grain separately versus a total mixed ration. J Dairy Sci 69:2140–2147. doi:10.3168/jds.S0022-0302(86)80646-4
Oude Elferinck SJ, Driehuis F, Becker PM, Gottschal JC, Faber F, Spoelstra SF (2001) The presence of Acetobacter sp. in ensiled forage crops and ensiled industrial byproducts. Med Fac Landbouwkd Univ Gent 66:427–430
Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML (2011) Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi:10.1128/AEM.02988-10
Welkie DG, Stevenson DM, Weimer PJ (2009) ARISA analysis of ruminal bacterial community dynamics in lactating dairy cows during the feeding cycle. Anaerobe 16:94–100. doi:10.1016/j.anaerobe.2009.07.002
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by JSPS KAKENHI Grant (Grant No. 24580390).
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Tang, M.T., Han, H., Yu, Z. et al. Variability, stability, and resilience of fecal microbiota in dairy cows fed whole crop corn silage. Appl Microbiol Biotechnol 101, 6355–6364 (2017). https://doi.org/10.1007/s00253-017-8348-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8348-8