Abstract
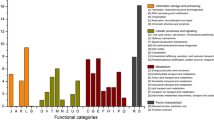
Although the taxonomical/phylogenetic diversity of microbial communities in biological heap leaching systems has been investigated, the diversity of functional genes was still unclear, and, especially, the differentiation and the relationships of diversity and functions of microbial communities in leaching heap (LH) and leaching solution (LS) were also still unclear. In our study, a functional gene array (GeoChip 5.0) was employed to investigate the functional gene diversity, and 16S rRNA gene sequencing was used to explore the taxonomical/phylogenetic diversity of microbial communities in LH and LS subsystems of Dexing copper mine (Jiangxi, China). Detrended correspondence analysis (DCA) showed that both functional gene structure and taxonomical/phylogenetic structure of microbial communities were significantly different between LH and LS. Signal intensities of genes, including genes for sulfur oxidation (e.g., soxB), metal homeostasis (e.g., arsm), carbon fixation (e.g., rubisco), polyphosphate degradation (e.g., ppk), and organic remediation (e.g., hydrocarbons) were significantly higher in LH, while signal intensities of genes for carbon degradation (e.g., amyA), polyphosphate synthesis (e.g., ppx), and sulfur reduction (e.g., dsrA) were significantly higher in LS. Further inspection revealed that microbial communities in LS and LH were dominated by Acidithiobacillus and Leptospirillum. However, rare species were relatively higher abundant in LH. Additionally, diversity index of functional genes was significantly different in LS (9.915 ± 0.074) and LH (9.781 ± 0.165), and the taxonomical/phylogenetic diversity index was also significantly different in LH (4.398 ± 0.508) and LS (3.014 ± 0.707). Functional tests, including sulfur-oxidizing ability, iron-oxidizing ability, and pyrite bioleaching ability, showed that all abilities of microbial communities were significantly stronger in LH than those in LS. Further studies found that most key genes (e.g., soxC and dsrA), rather than functional gene diversity index, were significantly correlated with abilities of microbial communities by linear regression analysis and Pearson correlation tests. In addition, the abilities were significantly correlated with taxonomical/phylogenetic diversity index and some rare species (e.g., Ferrithrix).





Similar content being viewed by others

References
Albuquerque L, Rainey FA, Nobre MF, Da CM (2012) Hydrotalea sandarakina sp. nov., isolated from a hot spring runoff, and emended descriptions of the genus Hydrotalea and the species Hydrotalea flava. Int J Syst Evol Micr 62:1603–1608. doi:10.1099/ijs.0.034496-0
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. doi:10.1016/S0168-6496(03)00028-X
Ben-Dov E, Brenner A, Kushmaro A (2007) Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microbial Ecol 54:439–451. doi:10.1007/s00248-007-9233-2
Borole AP, Neill HO, Tsouris C, Cesar S (2008) A microbial fuel cell operating at low pH using the acidophile Acidiphilium cryptum. Biotechnol Lett 30:1367–1372. doi:10.1007/s10529-008-9700-y
Bryant JA, Stewart FJ, Eppley JM, Delong EF (2012) Microbial community phylogenetic and trait diversity declines with depth in a marine oxygen minimum zone. Ecology 93:1659–1673. doi:10.1890/11-1204.1
Cong J, Liu X, Lu H, Xu H, Li Y, Deng Y, Li D, Zhang Y (2015) Analyses of the influencing factors of soil microbial functional gene diversity in tropical rainforest based on GeoChip 5.0. BMC Microbiol 15:397–398. doi:10.1016/j.gdata.2015.07.010
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 2004(14):927–930. doi:10.1111/j.1654-1103.2003.tb02228.x
Ehrlich HL (2001) Past, present and future of biohydrometallurgy. Hydrometallurgy 59:127–134. doi:10.1016/S0304-386X(00)00165-1
Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. P Natl Acad Sci USA 109:21390–21395. doi:10.1073/pnas.1215210110
Frediani M, Mezzanotte R, Vanni R, Pignone D, Cremonini R (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi:10.1038/nature02340
Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q, Hu Y, Li X, He Z, Ye D (2014) GeoChip as a metagenomics tool to analyze the microbial gene diversity along an elevation gradient. Genomics Data 2:132–134. doi:10.1016/j.gdata.2014.06.003
Guo X, Yin H, Cong J, Dai Z, Liang Y, Liu X (2013) RubisCO gene clusters found in a metagenome microarray from acid mine drainage. Appl Environ Microb 79:2019–2026. doi:10.1128/AEM.03400-12
Hu Q, Guo X, Liang Y, Hao X, Ma L, Yin H, Liu X (2015) Comparative metagenomics reveals microbial community differentiation in a biological heap leaching system. Res Microbiol 166:525–534. doi:10.1016/j.resmic.2015.06.005
Itoh T, Yoshikawa NT (2007) Thermogymnomonas acidicola gen. nov., sp nov., a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int J Syst Evol Micr 57:2557–2561. doi:10.1099/ijs.0.65203-0
Johnson DB (1998) Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol 27:307–317. doi:10.1016/S0168-6496(98)00079-8
Johnson DB, Bacelar-Nicolau P, Okibe N, Thomas A, Hallberg KB (2009) Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria. Int J Syst Evol Micr 59:1082–1089. doi:10.1099/ijs.0.65409-0
Joyeux C, Fouchard S, Llopiz P, Neunlist S (2004) Influence of the temperature and the growth phase on the hopanoids and fatty acids content of Frateuria aurantia (DSMZ 6220). FEMS Microbiol Ecol 47:371–379. doi:10.1016/S0168-6496(03)00302-7
Keshri J, Mankazana BBJ, Momba MNB (2015) Profile of bacterial communities in south African mine-water samples using Illumina next-generation sequencing platform. Appl Microbiol Biot 99:3233–3242. doi:10.1007/s00253-014-6213-6
Kuang J, Huang L, He Z, Chen L, Hua Z, Jia P, Li S, Liu J, Li J, Zhou J, Shu W (2016) Predicting taxonomic and functional structure of microbial communities in acid mine drainage. ISME J. doi:10.1038/ismej.2015.201
Liu H, Yin H, Dai Y, Dai Z, Yi L, Qian L, Jiang H, Liu X (2011) The co-culture of Acidithiobacillus ferrooxidans and Acidiphilium acidophilum enhances the growth, iron oxidation, and CO 2 fixation. Arch Microbiol 193:857–866. doi:10.1007/s00203-011-0723-8
Mendez-Garcia C, Pelaez AI, Mesa V, Sanchez J, Golyshina OV, Ferrer M (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475. doi:10.3389/fmicb.2015.00475
Osorio H, Martínez V, Veloso FA, Pedroso I, Valdés J, Jedlicki E, Holmes DS, Quatrini R (2008) Iron homeostasis strategies in acidophilic iron oxidizers: studies in Acidithiobacillus and Leptospirillum. Hydrometallurgy 94:175–179. doi:10.1016/j.hydromet.2008.05.038
Parton WJ, Stewart JWB, Cole CV (1988) Dynamics of C, N, P and S in grassland soils: a model. Biogeochemistry 5:109–131. doi:10.1007/BF02180320
Peres-Neto PR, Jackson DA (2001) How well do multivariate data sets match? The advantages of a procrustean superimposition approach over the mantel test. Oecologia 129:169–178. doi:10.1007/s004420100720
Pielou EC (1977) Mathematical ecology. J Anim Ecol 47
Rodrı́guez YA, Ballester ML, Blázquez FG, Muñoz JA (2003) New information on the chalcopyrite bioleaching mechanism at low and high temperature. Hydrometallurgy 71:47–56. doi:10.1016/S0304-386X(03)00173-7
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Slyemi D, Moinier D, Brochier-Armanet C, Bonnefoy V, Johnson DB (2011) Characteristics of a phylogenetically ambiguous, arsenic-oxidizing Thiomonas sp., Thiomonas arsenitoxydans strain 3As(T) sp nov. Arch Microbiol 193:439–449. doi:10.1007/s00203-011-0684-y
Smith TB, Wayne RK, Girman DJ, Bruford MW (1997) A role for ecotones in generating rainforest biodiversity. Science 276:1855–1857. doi:10.1126/science.276.5320.1855
Tu Q, Yu H, He Z, Deng Y, Wu L, Nostrand JDV, Zhou A, Voordeckers J, Lee YJ, Qin Y (2014) GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928. doi:10.1111/1755-0998.12239
Xiao Y, Xu YD, Dong W, Liang Y, Fan F, Zhang X, Zhang X, Niu J, Ma L, She S (2015) The complicated substrates enhance the microbial diversity and zinc leaching efficiency in sphalerite bioleaching system. Appl Microbiol Biot 99:10311–10322. doi:10.1007/s00253-015-6881-x
Xiao Y, Liu X, Ma L, Liang Y, Niu J, Gu Y, Zhang X, Hao X, Dong W, She S, Yin H (2016) Microbial communities from different subsystems in biological heap leaching system play different roles in iron and sulfur metabolisms. Appl Microbiol Biot. doi:10.1007/s00253-016-7537-1
Xie J, He Z, Liu X, Liu X, Van Nostrand JD, Deng Y, Wu L, Zhou J, Qiu G (2011) GeoChip-based analysis of the functional gene diversity and metabolic potential of microbial communities in acid mine drainage. Appl Environ Microb 77:991–999. doi:10.1128/AEM.01798-10
Yin H, Cao L, Qiu G, Wang D, Kellogg L, Zhou J, Dai Z, Liu X (2007) Development and evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in acid mine drainages and bioleaching systems. J Microbiol Meth 70:165–178. doi:10.1016/j.mimet.2007.04.011
Yin H, Zhang X, Li X, He Z, Liang Y, Guo X, Hu Q, Xiao Y, Cong J, Ma L (2014) Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol 14:1–14. doi:10.1186/1471-2180-14-179
Zhang X, Liu X, Liang Y, Fan F, Zhang X, Yin H (2016a) Metabolic diversity and adaptive mechanisms of iron- and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environ Microbiol Rep. doi:10.1111/1758-2229.12435
Zhang X, Niu J, Liang Y, Liu X, Yin H (2016b) Metagenome-scale analysis yields insights into the structure and function of microbial communities in a copper bioleaching heap. BMC Genet 17:1–12. doi:10.1186/s12863-016-0330-4
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict interests.
Electronic supplementary material
ESM 1
(PDF 495 kb)
Rights and permissions
About this article
Cite this article
Xiao, Y., Liu, X., Liang, Y. et al. Insights into functional genes and taxonomical/phylogenetic diversity of microbial communities in biological heap leaching system and their correlation with functions. Appl Microbiol Biotechnol 100, 9745–9756 (2016). https://doi.org/10.1007/s00253-016-7819-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7819-7