Abstract
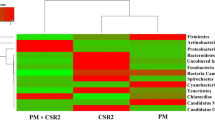
Silkworm (Bombyx mori L.) larvae were used as an ideal animal protein source for astronauts in the bioregenerative life support system (BLSS). Here, we compared the difference in bacterial communities of the silkworm larval gut between the BLSS rearing way (BRW) and the traditional rearing way (TRW) through culture-dependent approach, 16S rRNA gene analysis, and denaturing gradient gel electrophoresis (DGGE). The culture-dependent approach revealed that the numbers of gut bacteria of silkworm in the BRW significantly decreased compared with that of the TRW. The analysis of clone libraries showed that the gut microbiota in the BRW was significantly less diverse than that in the TRW. Acinetobacter and Bacteroides were dominant populations in the BRW, and Bacillus and Arcobacter dominated in the TRW. DGGE profiles confirmed the difference of silkworm gut bacterial community between two rearing ways. These results demonstrate that gut bacteria change from the BRW contributes to the decrease of silkworm physiological activity. This study increases our understanding of the change of silkworm gut microbiota in response to lettuce leaf feeding in the BRW. We could use the dominant populations to make probiotic products for nutrient absorption and disease prevention in the BLSS to improve gut microecology, as well as the yield and quality of animal protein.





Similar content being viewed by others
References
Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T, Geoffrey CJ, Vendan SE (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10:1–20
Ansari MA, Tirry L, Moens M (2003) Entomopathogenic nematodes and their symbiotic bacteria for the biological control of Hoplia philanthus (Coleoptera: Scarabaeidae). Biol Control 28:111–117
Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN (2012) Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e404017. doi:10.1371/journal.pone.004040
Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70:293–300
Butler JL, Williams MA, Bottomley PJ, Myrold DD (2003) Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol 69:6793–6800
Ceuppens S, Boon N, Uyttendaele M (2013) Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol Ecol 1:1–18
Dillon RJ, Vennard CT, Buckling A, Charnley AK (2005) Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–1298
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Kalpana S, Hatha AAM, Lakshmanaperumalsamy P (1994) Gut microflora of the larva of silkworm, Bombyx mori. Insect Sci Appl 15:499–502
Kiorboe T, Tang K, Grossart HP, Ploug H (2003) Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl Environ Microbiol 69:3036–3047
Lu X, Wang F (2002) Inhibition of cultured supernatant of Enterococci strains on germination of Nosema bombycis spores in vitro. Sci Sericulture 28:126–128 (In Chinese)
McKillip JL, Small CL, Brown JL, Brunner JF, Spence KD (1997) Sporogenous midgut bacteria of the leafroller, Pandemis pyrusana (Lepidoptera: Tortricidae). Environ Entomol 26:1475–1481
Militza CC, Nakatsu CH, Konopka A (2006) Effect of nutrient periodicity on microbial community dynamics. Appl Environ Microbiol 72:3175–3183
Nakatsu CH, Torsvikb V, Øvreås L (2000) Soil community analysis using DGGE of 16S rDNA polymerase chain reaction products. Soil Sci Soc AM J 64:1382–1388
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salam LB, Obayori OS, Olatoye NO (2013) Biodegradation of anthracene by a novel actinomycete, Microbacterium sp. isolated from tropical hydrocarbon-contaminated soil. World J Microbiol Biotechnol. doi:10.1007/s11274-013-1437-7
Schäfer A, Konrad R, Kuhnigk T, Kämpfer P, Hertel H, König H (1996) Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Bacteriol 5:471–478
Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF (2006) Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188:8272–8282
Shi W, Uzuner U, Huang L, Jesudhasan PR, Pillai SD, Yuan JS (2011) Comparative analysis of insect gut symbionts for composition–function relationships and biofuel application potential. Biofuels 5:529–544
Subramanian S, Gadhave KR, Mohanraj P, Thangamalar A (2009) Use of 16S rRNA probes for characterization of gut microflora of silkworm (Bombyx mori L.) breeds. Karnataka J Agric Sci 22:476–478
Takizawa Y, Iizuka T (1968) The aerobic bacterial flora in the gut of larvae of the silkworm Bombyx mori L. (I) The relation between media and the numbers of living cells. J Sericultural Sci Jpn 37:295–305
Tian Z, Hui F, Ke T, Kan Y, Wen Z (2007) Molecular analysis of the bacteria community composition in silkworm midgut. Sci Sericulture 33:592–595 (In Chinese)
Weber S, Stubner S, Conrad R (2001) Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl Environ Microbiol 67:1318–1327
Yang Y, Tang L, Tong L, Liu H (2009) Silkworms culture as a source of protein for humans in space. Adv Space Res 43:1236–1242
Xiang H, Li M, Zhao Y, Zhao L, Zhang Y, Huang Y (2007) Bacterial community in midguts of the silkworm larvae estimated by PCR\DGGE and 16SrDNA gene library analysis. Acta Entomol Sin 3:222–233
Yu X, Liu H, Tong L (2008) Feeding scenario of the silkworm (Bombyx mori L.) in the BLSS. Acta Astronaut 63:1086–1092
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012DFR30570) and National Natural Science Foundation of China (Grant No.31301706).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xue Liang and Yuming Fu contributed equally to this study.
Rights and permissions
About this article
Cite this article
Liang, X., Fu, Y., Tong, L. et al. Microbial shifts of the silkworm larval gut in response to lettuce leaf feeding. Appl Microbiol Biotechnol 98, 3769–3776 (2014). https://doi.org/10.1007/s00253-014-5532-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5532-y