Abstract
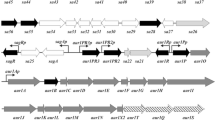
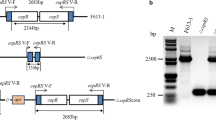
Clusters for clavulanic acid (CA) biosynthesis are present in the actinomycetes Streptomyces flavogriseus ATCC 33331 and Saccharomonospora viridis DSM 43017. These clusters, which are silent, contain blocks of conserved genes in the same order as those of the Streptomyces clavuligerus CA cluster but assembled in a different organization. S. flavogriseus was grown in nine different media, but clavulanic acid production was undetectable using bioassays or by high-performance liquid chromatography analyses. Reverse-transcriptase polymerase chain reaction (RT-PCR) of S. flavogriseus CA biosynthesis genes showed that the regulatory genes ccaR and claR and some biosynthetic genes were expressed whereas expression of cyp, orf12, orf13, and oppA2 was undetectable. The ccaR gene of S. clavuligerus was unable to switch on CA production in S. flavogriseus::[Pfur-ccaR C], but insertion of a cosmid carrying the S. clavuligerus CA cluster (not including the ccaR gene) conferred clavulanic acid production on S. flavogriseus::[SCos-CA] particularly in TBO and YEME media; these results suggests that some of the S. flavogriseus CA genes are inactive. The known heptameric sequences recognized by CcaR in S. clavuligerus are poorly or not conserved in S. flavogriseus. Quantitative RT-PCR analysis of the CA gene clusters of S. clavuligerus and S. flavogriseus showed that the average expression value of the expressed genes in the former strain was in the order of 1.68-fold higher than in the later. The absence of CA production by S. flavogriseus can be traced to the lack of expression of the essential genes cyp, orf12, orf13, orf14, and oppA2. Heterologous expression of S. clavuligerus CA gene cluster in S. flavogriseus::[SCos-CA] was 11- to 14-fold lower than in the parental strain, suggesting that the genetic background of the host strain is important for optimal production of CA in Streptomyces.





Similar content being viewed by others
References
Aidoo KA, Wong A, Alexander DC, Rittammer RA, Jensen SE (1994) Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene 147:41–46
Aigle B, Wietzorrek A, Takano E, Bibb MJ (2000) A single amino acid substitution in region 1.2 of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol Microbiol 37:995–1004
Alexander DC, Jensen SE (1998) Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J Bacteriol 180:4068–4079
Arulanantham H, Kershaw NJ, Hewitson KS, Hughes CE, Thirkettle JE, Schofield CJ (2006) ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. J Biol Chem 6:279–287
Bachmann BO, Li R, Townsend CA (1998) Beta-Lactam synthetase: a new biosynthetic enzyme. Proc Natl Acad Sci USA 95:9082–9086
Barreiro C, Prieto C, Sola-Landa A, Solera E, Martínez-Castro M, Pérez-Redondo R, García-Estrada C, Aparicio JF, Fernández-Martínez LT, Santos-Aberturas J, Salehi-Najafabadi Z, Rodríguez-García A, Tauch A, Martín JF (2012) Draft genome of Streptomyces tsukubaensis NRRL 18488, the producer of the clinically important immunosuppressant Tacrolimus (FK506). J Bacteriol 174:3756–3757
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bérdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are. J Antibiot 65:385–395
Bignell DR, Tahlan K, Colvin KR, Jensen SE, Leskiw BK (2005) Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob Agents Chemother 49:1529–1541
Blanco G (2012) Comparative analysis of a cryptic thienamycin-like gene cluster identified in Streptomyces flavogriseus by genome mining. Arch Microbiol 194:549–555
Brakhage AA, Schroeckh V (2011) Fungal secondary metabolites - strategies to activate silent gene clusters. Fungal Genet Biol 48:15–22
Doull JL, Vining LC (1989) Culture conditions promoting dispersed growth and biphasic production of actinorhodin in shaken cultures of Streptomyces coelicolor A3(2). FEMS Microbiol Lett 3:265–268
Enguita FJ, Coque JJ, Liras P, Martín JF (1998) The nine genes of the Nocardia lactamdurans cephamycin cluster are transcribed into large mRNAs from three promoters, two of them located in a bidirectional promoter region. J Bacteriol 180:5489–5494
Eustáquio AS, Gust B, Galm U, Li SM, Chater KF, Heide L (2005) Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters. Appl Environ Microbiol 71:2452–2459
Flinspach K, Westrich L, Kaysser L, Siebenberg S, Gomez-Escribano JP, Bibb M, Gust B, Heide L (2010) Heterologous expression of the biosynthetic gene clusters of coumermycin A(1), clorobiocin and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93:823–832
Foulstone M, Reading C (1982) Assay of amoxicillin and clavulanic acid, the components of Augmentin, in biological fluids with high-performance liquid chromatography. Antimicrob Agents Chemoter 22:753–762
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215
Gunsinor M, Breazeale SD, Lind AJ, Ravel J, Janc JW, Townsend CA (2004) The biosynthetic gene cluster for a monocyclic beta-lactam antibiotic, nocardicin A. Chem Biol 11:927–938
Higgens CE, Hamill RL, Sands TH, Hoehn MM, Davis NE, Najarahan R, Boeck LD (1974) The occurrence of desacetoxycephalosporin C in fungi and Streptomyces. J Antibiot 27:298–300
Huang S, Yudong Z, Zhiwei Q, Xiaoling W, Mayca O, Chen L, He J, Yu Y, Deng H (2011) Identification and heterologous expression of the biosynthetic gene cluster for holomycin produced by Streptomyces clavuligerus. Process Biochem 46:811–816
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Ishida K, Hung TV, Liou K, Lee HC, Shin CH, Sohng JK (2006) Characterization of pbpA and pbp2 encoding penicillin-binding proteins located on the downstream of clavulanic acid gene cluster in Streptomyces clavuligerus. Biotechnol Lett 28:409–417
Janc JW, Egan LA, Townsend CA (1995) Purification and characterization of clavaminate synthase from Streptomyces antibioticus. A multifunctional enzyme of clavam biosynthesis. J Biol Chem 270:5399–5404
Khaleeli N, Li R, Townsend CA (1999) Origin of the β-lactam carbons in clavulanic acid from an usual thiamine pyrophosphate-mediated reaction. J Am Chem Soc 121:9223–9224
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. John Innes Foundation, Norwich
Kurt A, Álvarez-Alvarez R, Liras P, Özcengiz G (2013) Role of the cmcH–ccaR intergenic region and ccaR overexpression in cephamycin C biosynthesis in Streptomyces clavuligerus. Applied Microbiol Biotechnol. doi: 10.1007/s00253-013-4721-4724
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA 108:6258–6263
Li R, Khaleeli N, Townsend CA (2000) Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Bacteriol 182:4087–4095
Liras P, Rodríguez-García A, Martín JF (1998) Evolution of the clusters of genes for β-lactam antibiotics: a model for evolutive combinatorial assembly of new β-lactams. Internat Microbiol 1:271–278
Liras P, Santamarta I, Pérez-Redondo R (2011) Clavulanic acid and clavams biosynthesis and regulation. In: Dyson P (ed) Streptomyces Molecular Biology and Biotechnology. Caister Academic Press, Norfolk, UK, pp 167–178
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 22DDCt Method. Methods 25:402–408
López-García MT, Santamarta I, Liras P (2010) Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology 156:2354–2365
Lorenzana LM, Pérez-Redondo R, Santamarta I, Martín JF, Liras P (2004) Two oligopeptide-permease-encoding genes in the clavulanic acid cluster of Streptomyces clavuligerus are essential for production of the beta-lactamase inhibitor. J Bacteriol 186:3431–3438
Marsh EN, Chang MD, Townsend CA (1992) Two isozymes of clavaminate synthase central to clavulanic acid formation: cloning and sequencing of both genes from Streptomyces clavuligerus. Biochemistry 31:12648–12657
Martín JF, Liras P (2010) Engineering of Regulatory Cascades and Networks Controlling Antibiotic Biosynthesis in Streptomyces. Current Op in Microbiol 13:263–273
Martín JF, Gutiérrez S, Aparicio JF (2000) Secondary metabolites. In: Lederberg J (ed) Encyclopedia of Microbiology, vol 4, 2nd edn. Academic Press, San Diego, pp 213–236
Matthew AG, Till R, Smith MCM (2003) Integration site for Streptomyces phage ΦBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323
Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Müller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RA, Breitling R, Takano E (2010) The sequence of a 1.8-mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol 2:212–224
Mellado E, Lorenzana LM, Rodríguez-Sáiz M, Díez B, Liras P, Barredo JL (2002) The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 148:1427–1438
Nicholson NH, Baggaley KH, Cassels R, Davison M, Elson SW, Fulston M, Tyler JW, Woroniecki ST (1994) Evidence that the intermediate biosynthetic precursor of clavulanic acid is its N-aldehyde analogue. J Chem Soc Chem Commun 1994:1281–1282
Nobary GS, Jensen SE (2012) A comparison of the clavam biosynthetic gene clusters in Streptomyces antibioticus Tü1718 and Streptomyces clavuligerus. Can J Microbiol 58:413–425
Ortiz de Orue Lucana D, Tröller M, Schrempf H (2003) Amino acid residues involved in reversible thiol formation and zinc ion binding in the Streptomyces reticuli redox regulator FurS. Mol Genet Genomics 268:618–627
Paradkar AS, Aidoo KA, Jensen SE (1998) A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol 27:831–843
Pati A, Sikorski J, Nolan M, Lapidus A, Copeland A, Glavina del Rio T, Lucas S, Chen F, Tice H, Pitluck S, Cheng JF, Chertkov O, Brettin T, Han C, Detter JC, Kuske C, Bruce D, Goodwin L, Chain P, D'haeseleer P, Chen A, Palaniappan K, Ivanova N, Mavromatis K, Mikhailova N, Rohde M, Tindall BJ, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP (2009) Complete genome sequence of Saccharomonospora viridis type strain (P101). Stand Genomic Sci 1:141–149
Pérez-Llarena F, Martín JF, Galleni M, Coque JJ, Fuente JL, Frère JM, Liras P (1997a) The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A beta-lactamase of low enzymatic activity. J Bacteriol 179:6035–6040
Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF (1997b) A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both ß-lactam compounds. J Bacteriol 179:2053–2059
Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P (1998) The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211:311–321
Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P (1999) Deletion of the pyc gene blocks clavulanic acid biosynthesis except in glycerol-containing medium: evidence for two different genes in formation of the C3 unit. J Bacteriol 181:6922–6928
Robles-Reglero V, Santamarta I, Álvarez-Álvarez R, Martín JF, Liras P (2013) Transcriptional analysis and proteomics of the holomycin gene cluster in overproducer mutants of Streptomyces clavuligerus. J Biotechnol 163(1):69–76
Rodríguez-García A, Ludovice M, Martín JF, Liras P (1997) Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol Microbiol 25:219–28
Romero J, Liras P, Martín JF (1984) Dissociation of cephamycin and clavulanic acid biosynthesis in Streptomyces clavuligerus. Appl Microbiol Biotechnol 20:318–325
Romero J, Liras P, Martín JF (1986) Utilization of ornithine and arginine as specific precursors of clavulanic acid. Appl Environ Microbiol 52:892–897
Santamarta I, López-García MT, Kurt A, Nárdiz N, Pérez-Redondo R, Álvarez-Álvarez R, Martín JF, Liras P (2011) Characterization of DNA-binding sequences for CcaR in the cephamycin–clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol 81:968–981
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81
Tong L, Yuanyuan D, Qiu C, Jingtao Z, Weiming Z, Kui H, Wenli L (2013) Cloning, characterization and heterologous expression of the indolocarbazole biosynthetic gene cluster from marine-derived Streptomyces sanyensis FMA. Mar Drugs 11:466–488
Valentine BP, Bailey CR, Doherty A, Morris J, Elson SW, Baggaley KH, Nicholson NH (1993) Evidence that arginine is a later metabolic intermediate than ornithine in the biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Chem Soc Chem Commun 1993:1210–1211
Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H (2009) Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A 106:8617–8622
Ward JM, Hodgson JE (1993) The biosynthetic genes for clavulanic acid and cephamycin production occur as a 'super-cluster' in three Streptomyces. FEMS Microbiol Lett 110:239–242
Xu G, Wang J, Wang L, Tian X, Yang H, Fan K, Yang K, Tan H (2010) "Pseudo" gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem 285:27440–27448
Young TS, Walsh CT (2011) Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci USA 108(32):13053–13058
Zhou Z, Gu J, Du YL, Li YQ, Wang Y (2011) The -omics Era- a toward a systems-level understanding of Streptomyces. Current Genomics 12:404–416
Acknowledgments
This work was supported by grants BIO2009-09820 and LE046A11-2 from the Spanish Ministry of Economy and Competitivity and the Junta de Castilla y León, respectively. R. Álvarez-Álvarez and Y. Martínez-Burgo received PFU fellowships from the Spanish Ministry of Education, Culture and Sports. We appreciate the collaboration of Dr. T. López-García in the design of oligonucleotides and RT-qPCR experiments and the reception of plasmid pFL1272 from Dr. F. Lombo.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 355 kb)
Rights and permissions
About this article
Cite this article
Álvarez-Álvarez, R., Martínez-Burgo, Y., Pérez-Redondo, R. et al. Expression of the endogenous and heterologous clavulanic acid cluster in Streptomyces flavogriseus: why a silent cluster is sleeping. Appl Microbiol Biotechnol 97, 9451–9463 (2013). https://doi.org/10.1007/s00253-013-5148-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5148-7